Pre Gene Modal
BGIBMGA012107
Annotation
THO_complex_subunit_1_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.747
Sequence
CDS
ATGTCGAGCAAACCAGGATTCGAAGAACTCCGCACGAAATACAAAGATGTACTTTCTAAAGCCTTTGCAACGAACAACATTGACCTCATCGATTCTTTTTCCAAAAACACTGACGAAACTGACAGAAAAGCAGCTATGGACCAAGCATTTAGGGATAAGCTACTTGAATTACTTACTGAGGATCATAACACTCTTGAAAATTTTGTATCGTTCTGCATAGAATCATGCAGGCGACTTATGGTTACGCCAACAATGCCTGTTGTATTACTTGGCGATATCTTTGACGCCCTCACACTAAATAAATGTGAACAATTATTCTCATACATTGAAAAAGGTGTCAGTATATGGAAGGAAGAACTATTTTTCGTGGCATGCAAAAATAATTTATTGAGAATGTGTAATGATTTATTAAGAAGACTTTCTCGATCTCAAAATACAGTTTTCTGTGGACGTATTCTACTTTTTTTGGCAAAATTCTTTCCATTCTCTGAACGCTCCGGTCTTAATATAGTCTCTGAATTCAATTTAGAAAATATAACGGAATTTGGTGTTGATAATTCAAGTACACTGAAAGATGCTTTGGAAGAAGAAATGGTTATTGATAATGAAAAACATAAACTGACTATAGATTACAATTTATACTGTAGATTCTGGAGTTTGCAAGAATTCTTCAGAAATCCTAATACTTGTTACAATAAACTCCAGTGGAAGACCTTTGTTGCACACTCTGGTAGTGTATTGTCCGCATTCTCATCCTACAAGCTAGAGACAGTTGAGCTGCAGAAATCTAAAATGAATCGATTGAAGCCATTGACTGACGTGGAAATGCAGGGTCAAGGCAAAGAGCAGCATTACTTTGCAAAATTTCTAACCAATCAGAAATTGTTAGAGTTGCAATTATCAGATTCGAACTTCAGAAGATGCGTCCTCATACAGTTTCTAATACTGTTTCAGTATCTGATGTCAACTGTTAAGTTCAAAACAGAAGCGCAGGAACTAAAATCTGACCAGTCTGATTGGATTAAAGATACAACCTCTCAGATTTATCGTTTGCTCGGTGAGACTCCACCGAATGGGAAGAAATTTGCGGAATGCGTTAAAAGCATACTAAAGCGAGAAGAACATTGGAACAACTGGAAAAATGATGGGTGTCCAGAATTTCAAAAACCCAAGCCACCGACACAATTGGACACCCCTGAAGATGTTAAGAAATCACGCAAAAGGCGTCGTCCGGTCGGCGATATTGTTAAGGAATACGAAGGACAGAAGAAATCCTATATGGGAAACAATGAGTTGACGAAACTCTGGAATCTGTGTCCGGATAATTTAGCGGCCTGTCGCACGAAGGAACGCGATTTCATGCCTTCGTTGGAGTCGTACATATCGAGCGCTTGTGACGAGTCGGGGCGGCGGCGCACGGCGGAGGGGGGTTGGGGGTGGCGCGCGCTGCGGCTCCTGGCGCGACGCTCGCCGCACTTCTTCGTGCAAACCAACAACCCTATAGGACGTCTCCCGGACTACCTCGATGCTATGGTCCATCGCATCACGAACGAGATGGCCGCCAACGCTGCGGCCGCCACCACCGAGCAGTCGCCGAAATCCGACGACAAGGCCAACAAGCCGGATGTGACCGAGGAGGAAATTAGTGAAGAACAAATGGAGGCTGATCTCATAAAAGAAGGCGACGCGAATGATATTGAACAGGTGCCCGACTCTACTCACGTCGGCGACGACGACTACGAGAAGTCGTCCAGGTCCCGCGTCACGATGATCACGCCCGCCCAGCTGGACGCGGTATCGGCCCAGCTCTCGGACTGGAAGACGCTGGCCGTGAAGCTCGGCTACAAGCCGGACGAGATCCAGTACTTCGAGACGGAAAACGCAACGGACGTCACGCGAGCCAAGAACATGCTTCAGCTCTGGTTCGACGACGACGAAGACGCGTCGGTCGAGAACCTCCTCTACACTATGGAGGGTCTGAAGATGACCGAGGCCTGCGAGGCGCTCAGACGAGTTATTTGA
Protein
MSSKPGFEELRTKYKDVLSKAFATNNIDLIDSFSKNTDETDRKAAMDQAFRDKLLELLTEDHNTLENFVSFCIESCRRLMVTPTMPVVLLGDIFDALTLNKCEQLFSYIEKGVSIWKEELFFVACKNNLLRMCNDLLRRLSRSQNTVFCGRILLFLAKFFPFSERSGLNIVSEFNLENITEFGVDNSSTLKDALEEEMVIDNEKHKLTIDYNLYCRFWSLQEFFRNPNTCYNKLQWKTFVAHSGSVLSAFSSYKLETVELQKSKMNRLKPLTDVEMQGQGKEQHYFAKFLTNQKLLELQLSDSNFRRCVLIQFLILFQYLMSTVKFKTEAQELKSDQSDWIKDTTSQIYRLLGETPPNGKKFAECVKSILKREEHWNNWKNDGCPEFQKPKPPTQLDTPEDVKKSRKRRRPVGDIVKEYEGQKKSYMGNNELTKLWNLCPDNLAACRTKERDFMPSLESYISSACDESGRRRTAEGGWGWRALRLLARRSPHFFVQTNNPIGRLPDYLDAMVHRITNEMAANAAAATTEQSPKSDDKANKPDVTEEEISEEQMEADLIKEGDANDIEQVPDSTHVGDDDYEKSSRSRVTMITPAQLDAVSAQLSDWKTLAVKLGYKPDEIQYFETENATDVTRAKNMLQLWFDDDEDASVENLLYTMEGLKMTEACEALRRVI
Summary
Uniprot
H9JRE7
A0A2H1WM04
A0A194Q9D2
A0A3S2NWG7
A0A194R543
A0A2A4JXE8
+ More
E9IQA4 A0A067RKL8 A0A088A7D4 A0A2J7RQX8 A0A0L7QUP7 A0A195BPU1 A0A154PRQ1 A0A0J7NZJ8 A0A195CWM9 F4X518 A0A151WYJ9 A0A2A3EB56 A0A310SEE7 E2ANQ3 A0A195DPJ3 A0A1W4WQ11 A0A195EYR7 E2B554 A0A158P2X1 A0A026WG44 A0A3L8DFV4 A0A1B0CCR7 A0A336M1X6 A0A1S4F250 A0A182YF18 A0A0M9A0A2 A0A1B0DDR4 A0A182NG63 A0A336KG73 Q17HT6 A0A0T6BD55 A0A182T6C0 A0A182JG52 A0A182QHV0 A0A182R3P9 A0A182TDI5 A0A182H6U2 A0A182K479 A0A182L7N9 A0A182UMF8 K7ISJ6 D6WX23 A0A182I826 A0A182MRI3 Q7QH59 A0A2P8Y2N2 A0A1I8JW94 A0A182VX40 A0A232FHI9 A0A182FQ83 A0A182PNK1 A0A2M4AGE0 A0A1B6EI84 A0A2M4AGF8 A0A0M8ZX54 W5JFR3 A0A1B6IAJ6 A0A1Y1MI33 A0A084VER7 A0A1I8NGH7 A0A1I8PR36 B0WA57 U4UCW0 N6UGB5 A0A2J7RQW9 A0A0L0CES3 B4JU04 T1PCA5 A0A034VHD5 A0A0K8UTJ7 B4LXR8 A0A0P5S7T9 A0A1B6CXE8 A0A1B0FAL1 A0A1A9Y8L4 A0A1A9VLK4 A0A1B0BPM3 A0A0P6G8W1 A0A240SX73 A0A0P5DLX7 A0A0N8BK75 A0A0P6EVF8 A0A0P5U6Y0 A0A0P5VPQ1 A0A1A9X2K4 A0A0P4Z9Y4 A0A0N8EN76 A0A0P5A5H4 A0A0P5NQP9 A0A1B6JPT5 A0A0P5V3Q0 A0A0P5R177
E9IQA4 A0A067RKL8 A0A088A7D4 A0A2J7RQX8 A0A0L7QUP7 A0A195BPU1 A0A154PRQ1 A0A0J7NZJ8 A0A195CWM9 F4X518 A0A151WYJ9 A0A2A3EB56 A0A310SEE7 E2ANQ3 A0A195DPJ3 A0A1W4WQ11 A0A195EYR7 E2B554 A0A158P2X1 A0A026WG44 A0A3L8DFV4 A0A1B0CCR7 A0A336M1X6 A0A1S4F250 A0A182YF18 A0A0M9A0A2 A0A1B0DDR4 A0A182NG63 A0A336KG73 Q17HT6 A0A0T6BD55 A0A182T6C0 A0A182JG52 A0A182QHV0 A0A182R3P9 A0A182TDI5 A0A182H6U2 A0A182K479 A0A182L7N9 A0A182UMF8 K7ISJ6 D6WX23 A0A182I826 A0A182MRI3 Q7QH59 A0A2P8Y2N2 A0A1I8JW94 A0A182VX40 A0A232FHI9 A0A182FQ83 A0A182PNK1 A0A2M4AGE0 A0A1B6EI84 A0A2M4AGF8 A0A0M8ZX54 W5JFR3 A0A1B6IAJ6 A0A1Y1MI33 A0A084VER7 A0A1I8NGH7 A0A1I8PR36 B0WA57 U4UCW0 N6UGB5 A0A2J7RQW9 A0A0L0CES3 B4JU04 T1PCA5 A0A034VHD5 A0A0K8UTJ7 B4LXR8 A0A0P5S7T9 A0A1B6CXE8 A0A1B0FAL1 A0A1A9Y8L4 A0A1A9VLK4 A0A1B0BPM3 A0A0P6G8W1 A0A240SX73 A0A0P5DLX7 A0A0N8BK75 A0A0P6EVF8 A0A0P5U6Y0 A0A0P5VPQ1 A0A1A9X2K4 A0A0P4Z9Y4 A0A0N8EN76 A0A0P5A5H4 A0A0P5NQP9 A0A1B6JPT5 A0A0P5V3Q0 A0A0P5R177
Pubmed
EMBL
BABH01016296
BABH01016297
ODYU01009489
SOQ53962.1
KQ459252
KPJ02108.1
+ More
RSAL01000045 RVE50609.1 KQ460779 KPJ12360.1 NWSH01000397 PCG76691.1 GL764688 EFZ17246.1 KK852621 KDR20035.1 NEVH01000613 PNF43227.1 KQ414734 KOC62348.1 KQ976432 KYM87514.1 KQ435012 KZC13790.1 LBMM01000669 KMQ97825.1 KQ977185 KYN05086.1 GL888680 EGI58475.1 KQ982649 KYQ52964.1 KZ288296 PBC28955.1 KQ770367 OAD52665.1 GL441295 EFN64939.1 KQ980713 KYN14404.1 KQ981905 KYN33440.1 GL445712 EFN89189.1 ADTU01007640 KK107231 EZA55022.1 QOIP01000009 RLU19053.1 AJWK01007043 UFQT01000289 SSX22833.1 KQ435782 KOX74745.1 AJVK01032172 AJVK01032173 UFQS01000316 UFQT01000316 SSX02778.1 SSX23149.1 CH477245 EAT46256.1 LJIG01001676 KRT85264.1 AXCN02001190 JXUM01027326 KQ560786 KXJ80892.1 KQ971361 EFA08780.1 APCN01002609 AXCM01006183 AAAB01008817 EAA05359.4 PYGN01001007 PSN38522.1 NNAY01000206 OXU30033.1 GGFK01006518 MBW39839.1 GECZ01032136 JAS37633.1 GGFK01006543 MBW39864.1 KQ435811 KOX72920.1 ADMH02001627 ETN61725.1 GECU01034797 GECU01023769 JAS72909.1 JAS83937.1 GEZM01030794 GEZM01030793 GEZM01030792 GEZM01030791 GEZM01030790 GEZM01030789 JAV85323.1 ATLV01012288 KE524778 KFB36461.1 DS231869 EDS40886.1 KB632061 ERL88421.1 APGK01036278 KB740939 ENN77692.1 PNF43229.1 JRES01000501 KNC30755.1 CH916374 EDV91583.1 KA646319 AFP60948.1 GAKP01017028 GAKP01017026 GAKP01017024 JAC41924.1 GDHF01022307 JAI30007.1 CH940650 EDW67877.1 KRF83547.1 GDIQ01095527 JAL56199.1 GEDC01019164 GEDC01001624 JAS18134.1 JAS35674.1 CCAG010010971 JXJN01018092 GDIQ01036919 JAN57818.1 GDIP01154232 JAJ69170.1 GDIQ01169659 JAK82066.1 GDIQ01070303 JAN24434.1 GDIP01134544 GDIP01118982 GDIP01102686 LRGB01001801 JAL84732.1 KZS10424.1 GDIP01097190 JAM06525.1 GDIP01216267 JAJ07135.1 GDIQ01010260 JAN84477.1 GDIP01203783 JAJ19619.1 GDIQ01145730 JAL05996.1 GECU01006813 JAT00894.1 GDIP01120987 JAL82727.1 GDIQ01106936 JAL44790.1
RSAL01000045 RVE50609.1 KQ460779 KPJ12360.1 NWSH01000397 PCG76691.1 GL764688 EFZ17246.1 KK852621 KDR20035.1 NEVH01000613 PNF43227.1 KQ414734 KOC62348.1 KQ976432 KYM87514.1 KQ435012 KZC13790.1 LBMM01000669 KMQ97825.1 KQ977185 KYN05086.1 GL888680 EGI58475.1 KQ982649 KYQ52964.1 KZ288296 PBC28955.1 KQ770367 OAD52665.1 GL441295 EFN64939.1 KQ980713 KYN14404.1 KQ981905 KYN33440.1 GL445712 EFN89189.1 ADTU01007640 KK107231 EZA55022.1 QOIP01000009 RLU19053.1 AJWK01007043 UFQT01000289 SSX22833.1 KQ435782 KOX74745.1 AJVK01032172 AJVK01032173 UFQS01000316 UFQT01000316 SSX02778.1 SSX23149.1 CH477245 EAT46256.1 LJIG01001676 KRT85264.1 AXCN02001190 JXUM01027326 KQ560786 KXJ80892.1 KQ971361 EFA08780.1 APCN01002609 AXCM01006183 AAAB01008817 EAA05359.4 PYGN01001007 PSN38522.1 NNAY01000206 OXU30033.1 GGFK01006518 MBW39839.1 GECZ01032136 JAS37633.1 GGFK01006543 MBW39864.1 KQ435811 KOX72920.1 ADMH02001627 ETN61725.1 GECU01034797 GECU01023769 JAS72909.1 JAS83937.1 GEZM01030794 GEZM01030793 GEZM01030792 GEZM01030791 GEZM01030790 GEZM01030789 JAV85323.1 ATLV01012288 KE524778 KFB36461.1 DS231869 EDS40886.1 KB632061 ERL88421.1 APGK01036278 KB740939 ENN77692.1 PNF43229.1 JRES01000501 KNC30755.1 CH916374 EDV91583.1 KA646319 AFP60948.1 GAKP01017028 GAKP01017026 GAKP01017024 JAC41924.1 GDHF01022307 JAI30007.1 CH940650 EDW67877.1 KRF83547.1 GDIQ01095527 JAL56199.1 GEDC01019164 GEDC01001624 JAS18134.1 JAS35674.1 CCAG010010971 JXJN01018092 GDIQ01036919 JAN57818.1 GDIP01154232 JAJ69170.1 GDIQ01169659 JAK82066.1 GDIQ01070303 JAN24434.1 GDIP01134544 GDIP01118982 GDIP01102686 LRGB01001801 JAL84732.1 KZS10424.1 GDIP01097190 JAM06525.1 GDIP01216267 JAJ07135.1 GDIQ01010260 JAN84477.1 GDIP01203783 JAJ19619.1 GDIQ01145730 JAL05996.1 GECU01006813 JAT00894.1 GDIP01120987 JAL82727.1 GDIQ01106936 JAL44790.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000218220
UP000027135
+ More
UP000005203 UP000235965 UP000053825 UP000078540 UP000076502 UP000036403 UP000078542 UP000007755 UP000075809 UP000242457 UP000000311 UP000078492 UP000192223 UP000078541 UP000008237 UP000005205 UP000053097 UP000279307 UP000092461 UP000076408 UP000053105 UP000092462 UP000075884 UP000008820 UP000075901 UP000075880 UP000075886 UP000075900 UP000075902 UP000069940 UP000249989 UP000075881 UP000075882 UP000075903 UP000002358 UP000007266 UP000075840 UP000075883 UP000007062 UP000245037 UP000076407 UP000075920 UP000215335 UP000069272 UP000075885 UP000000673 UP000030765 UP000095301 UP000095300 UP000002320 UP000030742 UP000019118 UP000037069 UP000001070 UP000008792 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000076858 UP000091820
UP000005203 UP000235965 UP000053825 UP000078540 UP000076502 UP000036403 UP000078542 UP000007755 UP000075809 UP000242457 UP000000311 UP000078492 UP000192223 UP000078541 UP000008237 UP000005205 UP000053097 UP000279307 UP000092461 UP000076408 UP000053105 UP000092462 UP000075884 UP000008820 UP000075901 UP000075880 UP000075886 UP000075900 UP000075902 UP000069940 UP000249989 UP000075881 UP000075882 UP000075903 UP000002358 UP000007266 UP000075840 UP000075883 UP000007062 UP000245037 UP000076407 UP000075920 UP000215335 UP000069272 UP000075885 UP000000673 UP000030765 UP000095301 UP000095300 UP000002320 UP000030742 UP000019118 UP000037069 UP000001070 UP000008792 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000076858 UP000091820
PRIDE
SUPFAM
SSF47986
SSF47986
ProteinModelPortal
H9JRE7
A0A2H1WM04
A0A194Q9D2
A0A3S2NWG7
A0A194R543
A0A2A4JXE8
+ More
E9IQA4 A0A067RKL8 A0A088A7D4 A0A2J7RQX8 A0A0L7QUP7 A0A195BPU1 A0A154PRQ1 A0A0J7NZJ8 A0A195CWM9 F4X518 A0A151WYJ9 A0A2A3EB56 A0A310SEE7 E2ANQ3 A0A195DPJ3 A0A1W4WQ11 A0A195EYR7 E2B554 A0A158P2X1 A0A026WG44 A0A3L8DFV4 A0A1B0CCR7 A0A336M1X6 A0A1S4F250 A0A182YF18 A0A0M9A0A2 A0A1B0DDR4 A0A182NG63 A0A336KG73 Q17HT6 A0A0T6BD55 A0A182T6C0 A0A182JG52 A0A182QHV0 A0A182R3P9 A0A182TDI5 A0A182H6U2 A0A182K479 A0A182L7N9 A0A182UMF8 K7ISJ6 D6WX23 A0A182I826 A0A182MRI3 Q7QH59 A0A2P8Y2N2 A0A1I8JW94 A0A182VX40 A0A232FHI9 A0A182FQ83 A0A182PNK1 A0A2M4AGE0 A0A1B6EI84 A0A2M4AGF8 A0A0M8ZX54 W5JFR3 A0A1B6IAJ6 A0A1Y1MI33 A0A084VER7 A0A1I8NGH7 A0A1I8PR36 B0WA57 U4UCW0 N6UGB5 A0A2J7RQW9 A0A0L0CES3 B4JU04 T1PCA5 A0A034VHD5 A0A0K8UTJ7 B4LXR8 A0A0P5S7T9 A0A1B6CXE8 A0A1B0FAL1 A0A1A9Y8L4 A0A1A9VLK4 A0A1B0BPM3 A0A0P6G8W1 A0A240SX73 A0A0P5DLX7 A0A0N8BK75 A0A0P6EVF8 A0A0P5U6Y0 A0A0P5VPQ1 A0A1A9X2K4 A0A0P4Z9Y4 A0A0N8EN76 A0A0P5A5H4 A0A0P5NQP9 A0A1B6JPT5 A0A0P5V3Q0 A0A0P5R177
E9IQA4 A0A067RKL8 A0A088A7D4 A0A2J7RQX8 A0A0L7QUP7 A0A195BPU1 A0A154PRQ1 A0A0J7NZJ8 A0A195CWM9 F4X518 A0A151WYJ9 A0A2A3EB56 A0A310SEE7 E2ANQ3 A0A195DPJ3 A0A1W4WQ11 A0A195EYR7 E2B554 A0A158P2X1 A0A026WG44 A0A3L8DFV4 A0A1B0CCR7 A0A336M1X6 A0A1S4F250 A0A182YF18 A0A0M9A0A2 A0A1B0DDR4 A0A182NG63 A0A336KG73 Q17HT6 A0A0T6BD55 A0A182T6C0 A0A182JG52 A0A182QHV0 A0A182R3P9 A0A182TDI5 A0A182H6U2 A0A182K479 A0A182L7N9 A0A182UMF8 K7ISJ6 D6WX23 A0A182I826 A0A182MRI3 Q7QH59 A0A2P8Y2N2 A0A1I8JW94 A0A182VX40 A0A232FHI9 A0A182FQ83 A0A182PNK1 A0A2M4AGE0 A0A1B6EI84 A0A2M4AGF8 A0A0M8ZX54 W5JFR3 A0A1B6IAJ6 A0A1Y1MI33 A0A084VER7 A0A1I8NGH7 A0A1I8PR36 B0WA57 U4UCW0 N6UGB5 A0A2J7RQW9 A0A0L0CES3 B4JU04 T1PCA5 A0A034VHD5 A0A0K8UTJ7 B4LXR8 A0A0P5S7T9 A0A1B6CXE8 A0A1B0FAL1 A0A1A9Y8L4 A0A1A9VLK4 A0A1B0BPM3 A0A0P6G8W1 A0A240SX73 A0A0P5DLX7 A0A0N8BK75 A0A0P6EVF8 A0A0P5U6Y0 A0A0P5VPQ1 A0A1A9X2K4 A0A0P4Z9Y4 A0A0N8EN76 A0A0P5A5H4 A0A0P5NQP9 A0A1B6JPT5 A0A0P5V3Q0 A0A0P5R177
Ontologies
PATHWAY
GO
PANTHER
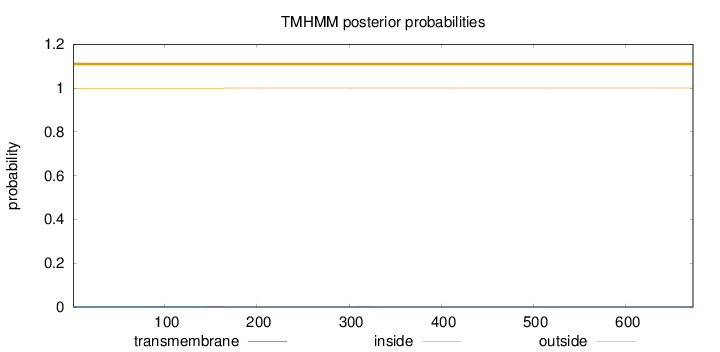
Topology
Length:
673
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01764
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00080
outside
1 - 673
Population Genetic Test Statistics
Pi
16.724903
Theta
17.411048
Tajima's D
0
CLR
14.282458
CSRT
0.370031498425079
Interpretation
Uncertain
