Gene
KWMTBOMO06799
Pre Gene Modal
BGIBMGA012044
Annotation
PREDICTED:_protein_FAM166B-like_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.647
Sequence
CDS
ATGTGGGTAGATGTGAGTGGGCGCGAAAAGCACAACTACATGACGCAGACAAATGGATGTTTTATACCGGGATACACTGGTCACTGCCCCATGTTGAAGTTTCGTTACGGAAAATGCTACGGTGATAATACGCGACAAATTCTACGTGAAATTCGAACCAAAGGACTTTTCAATAAACCATTCCAATACCGCACCGGCGACCATTACGAGTTGAACCAATTACCGCGACACGACGCTCCGCAACGAGATACCTACGACGGAATAGGAAACCGTCAAACCTCACACGTGACTGGATACACTGGATACGTACCCGGAATGAATTTTACTTACGGCAAGTCATATGGACGAACAGCGGACGATTGCATGGAAAACTTCGTGGACAACCAACGTGAATTACGTCGTAAAACTGATTTAAACAGAAGCTATATTAGATCGAGAAGTGCACCGAAAATGGAAACTGTACATTCTAGGGACGAAATACGGAGGGATCTCAGTAGATTCCGGGAGATTAATAAATATAAAGAAAACACAATATCGCCTGAGTTTCCACCAATAGCTGGCTACACGGGCCATATACCACGCATCAAGGGATCGGAAGCATCGTTGAGCCAAAGATACCATTGCGCTGCCAAACGGGGCCTTGAACTTATTAGACAGGAACGAGACACAAGAAAGGAACTCATAAACGCCGACACGAAAATTAGAACTATACTCAAAGATCATGATGATAAAAAATATTCATACTGGAACTGGGGATAG
Protein
MWVDVSGREKHNYMTQTNGCFIPGYTGHCPMLKFRYGKCYGDNTRQILREIRTKGLFNKPFQYRTGDHYELNQLPRHDAPQRDTYDGIGNRQTSHVTGYTGYVPGMNFTYGKSYGRTADDCMENFVDNQRELRRKTDLNRSYIRSRSAPKMETVHSRDEIRRDLSRFREINKYKENTISPEFPPIAGYTGHIPRIKGSEASLSQRYHCAAKRGLELIRQERDTRKELINADTKIRTILKDHDDKKYSYWNWG
Summary
Uniprot
A0A2A4JY48
A0A194Q9N8
A0A212EQ83
A0A194QPR0
A0A2H1WZ27
A0A0L7L1A8
+ More
A0A1Y1LJW9 A0A1Y1LH76 A0A139WLN5 A0A067RIT7 A0A1B6L935 A0A1B6M8P7 A0A1B6FUA9 A0A1Y1LIQ8 A0A1Y1LKX4 A0A1Y1LF05 A0A1Y1LH93 A0A2J7RSD9 A0A1B6IB31 E0V9S7 J9LV09 A0A2S2PYG5 A0A1Y1LF43 K7JQ24 A0A232FI72 A0A2A3ES11 A0A088AP87 A0A0J7LAD2 A0A0L7R0H4 A0A195B990 A0A158NL07 A0A195CNS6 A0A151X443 E2ALC4 A0A026WMJ2 F4WB69 A0A154PP91 E2C2X1 A0A0N0U5M9 A0A1S3IG61 A0A210R5D7 K1PU44 A0A0B6YVV5 V4BUX1 C1LFI5 A0A2T7NL09 A0A3M7SU91 A0A075A3B0 A0A177B0G0 H2KV50 Q86DY4 A0A3Q0KKG8 A0A183SZ81 A0A0X3PDQ0 A0A0L8H118 R7UKN5 G4LVN1 B3GUX2 A0A3P7LFY0 A0A2H1C4H5 A0A1S8WLK8 A0A183KFZ5 A0A1I8FW21 A0A1I8FZ71 T1F907 A0A094ZT67 A0A183VVR9 A0A183APF9 M3XJG1 F1SG06 M3WZI7 A0A341CVQ6 A0A0D9RD84 A0A2Y9J762 A0A3Q7X6D7 A0A3Q7XHU8 A7RIM2 A0A3Q7WB28 M3Y1F1 G1LS70 D2HAK2 A0A341CY54 A0A2U4CG61 A0A3Q0E8Y2 A0A1A6HZ27 A0A2Y9QKB1 A0A1U7R146 A0A212EMF4
A0A1Y1LJW9 A0A1Y1LH76 A0A139WLN5 A0A067RIT7 A0A1B6L935 A0A1B6M8P7 A0A1B6FUA9 A0A1Y1LIQ8 A0A1Y1LKX4 A0A1Y1LF05 A0A1Y1LH93 A0A2J7RSD9 A0A1B6IB31 E0V9S7 J9LV09 A0A2S2PYG5 A0A1Y1LF43 K7JQ24 A0A232FI72 A0A2A3ES11 A0A088AP87 A0A0J7LAD2 A0A0L7R0H4 A0A195B990 A0A158NL07 A0A195CNS6 A0A151X443 E2ALC4 A0A026WMJ2 F4WB69 A0A154PP91 E2C2X1 A0A0N0U5M9 A0A1S3IG61 A0A210R5D7 K1PU44 A0A0B6YVV5 V4BUX1 C1LFI5 A0A2T7NL09 A0A3M7SU91 A0A075A3B0 A0A177B0G0 H2KV50 Q86DY4 A0A3Q0KKG8 A0A183SZ81 A0A0X3PDQ0 A0A0L8H118 R7UKN5 G4LVN1 B3GUX2 A0A3P7LFY0 A0A2H1C4H5 A0A1S8WLK8 A0A183KFZ5 A0A1I8FW21 A0A1I8FZ71 T1F907 A0A094ZT67 A0A183VVR9 A0A183APF9 M3XJG1 F1SG06 M3WZI7 A0A341CVQ6 A0A0D9RD84 A0A2Y9J762 A0A3Q7X6D7 A0A3Q7XHU8 A7RIM2 A0A3Q7WB28 M3Y1F1 G1LS70 D2HAK2 A0A341CY54 A0A2U4CG61 A0A3Q0E8Y2 A0A1A6HZ27 A0A2Y9QKB1 A0A1U7R146 A0A212EMF4
Pubmed
EMBL
NWSH01000397
PCG76696.1
KQ459252
KPJ02124.1
AGBW02013309
OWR43621.1
+ More
KQ461185 KPJ07492.1 ODYU01012169 SOQ58340.1 JTDY01003743 KOB69059.1 GEZM01057595 JAV72235.1 GEZM01057588 JAV72241.1 KQ971319 KYB28816.1 KK852480 KDR22948.1 GEBQ01019856 JAT20121.1 GEBQ01007696 JAT32281.1 GECZ01015963 JAS53806.1 GEZM01057589 JAV72240.1 GEZM01057594 JAV72236.1 GEZM01057587 JAV72242.1 GEZM01057591 JAV72238.1 NEVH01000260 PNF43756.1 GECU01023560 JAS84146.1 DS234996 EEB10133.1 ABLF02039802 ABLF02039813 ABLF02039824 GGMS01001288 MBY70491.1 GEZM01057586 JAV72243.1 AAZX01002692 NNAY01000158 OXU30446.1 KZ288193 PBC34062.1 LBMM01000136 KMR04798.1 KQ414670 KOC64345.1 KQ976556 KYM80797.1 ADTU01019091 ADTU01019092 ADTU01019093 KQ977532 KYN02127.1 KQ982557 KYQ55044.1 GL440589 EFN65771.1 KK107152 QOIP01000007 EZA57128.1 RLU20055.1 GL888057 EGI68576.1 KQ435007 KZC13689.1 GL452255 EFN77675.1 KQ435783 KOX74671.1 NEDP02000337 OWF56104.1 JH817628 EKC27817.1 HACG01013021 CEK59886.1 KB202014 ESO92829.1 FN317733 CAX73463.1 PZQS01000011 PVD21849.1 REGN01000752 RNA39383.1 KL596631 KER32727.1 LWCA01000617 OAF67610.1 DF144317 GAA31536.2 AY223437 AAP06473.1 UYSU01035298 VDL95914.1 GEEE01013144 JAP50081.1 KQ419615 KOF82887.1 AMQN01007192 KB300180 ELU07069.1 CABG01000016 CCD58970.1 EZ000118 ACE06898.1 UYRU01063722 VDN15815.1 KZ431115 PIS82368.1 KV905557 OON15348.1 UZAK01036279 VDP54688.1 NIVC01005038 PAA46132.1 NIVC01003104 NIVC01000434 PAA53303.1 PAA83063.1 AMQM01005170 KB096830 ESO01184.1 KL250758 KGB36269.1 UZAO01052872 VDQ00455.1 UZAN01046551 VDP84292.1 AFYH01120918 AFYH01120919 AFYH01120920 AFYH01120921 AEMK02000005 DQIR01029370 HCZ84845.1 AANG04001372 AQIB01022057 DS469512 EDO48739.1 AEYP01022147 ACTA01186951 GL192634 EFB15365.1 LZPO01005771 OBS83491.1 AGBW02013840 OWR42672.1
KQ461185 KPJ07492.1 ODYU01012169 SOQ58340.1 JTDY01003743 KOB69059.1 GEZM01057595 JAV72235.1 GEZM01057588 JAV72241.1 KQ971319 KYB28816.1 KK852480 KDR22948.1 GEBQ01019856 JAT20121.1 GEBQ01007696 JAT32281.1 GECZ01015963 JAS53806.1 GEZM01057589 JAV72240.1 GEZM01057594 JAV72236.1 GEZM01057587 JAV72242.1 GEZM01057591 JAV72238.1 NEVH01000260 PNF43756.1 GECU01023560 JAS84146.1 DS234996 EEB10133.1 ABLF02039802 ABLF02039813 ABLF02039824 GGMS01001288 MBY70491.1 GEZM01057586 JAV72243.1 AAZX01002692 NNAY01000158 OXU30446.1 KZ288193 PBC34062.1 LBMM01000136 KMR04798.1 KQ414670 KOC64345.1 KQ976556 KYM80797.1 ADTU01019091 ADTU01019092 ADTU01019093 KQ977532 KYN02127.1 KQ982557 KYQ55044.1 GL440589 EFN65771.1 KK107152 QOIP01000007 EZA57128.1 RLU20055.1 GL888057 EGI68576.1 KQ435007 KZC13689.1 GL452255 EFN77675.1 KQ435783 KOX74671.1 NEDP02000337 OWF56104.1 JH817628 EKC27817.1 HACG01013021 CEK59886.1 KB202014 ESO92829.1 FN317733 CAX73463.1 PZQS01000011 PVD21849.1 REGN01000752 RNA39383.1 KL596631 KER32727.1 LWCA01000617 OAF67610.1 DF144317 GAA31536.2 AY223437 AAP06473.1 UYSU01035298 VDL95914.1 GEEE01013144 JAP50081.1 KQ419615 KOF82887.1 AMQN01007192 KB300180 ELU07069.1 CABG01000016 CCD58970.1 EZ000118 ACE06898.1 UYRU01063722 VDN15815.1 KZ431115 PIS82368.1 KV905557 OON15348.1 UZAK01036279 VDP54688.1 NIVC01005038 PAA46132.1 NIVC01003104 NIVC01000434 PAA53303.1 PAA83063.1 AMQM01005170 KB096830 ESO01184.1 KL250758 KGB36269.1 UZAO01052872 VDQ00455.1 UZAN01046551 VDP84292.1 AFYH01120918 AFYH01120919 AFYH01120920 AFYH01120921 AEMK02000005 DQIR01029370 HCZ84845.1 AANG04001372 AQIB01022057 DS469512 EDO48739.1 AEYP01022147 ACTA01186951 GL192634 EFB15365.1 LZPO01005771 OBS83491.1 AGBW02013840 OWR42672.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
UP000007266
+ More
UP000027135 UP000235965 UP000009046 UP000007819 UP000002358 UP000215335 UP000242457 UP000005203 UP000036403 UP000053825 UP000078540 UP000005205 UP000078542 UP000075809 UP000000311 UP000053097 UP000279307 UP000007755 UP000076502 UP000008237 UP000053105 UP000085678 UP000242188 UP000005408 UP000030746 UP000245119 UP000276133 UP000008854 UP000050788 UP000275846 UP000053454 UP000014760 UP000050789 UP000095280 UP000215902 UP000015101 UP000050795 UP000050740 UP000008672 UP000008227 UP000011712 UP000252040 UP000029965 UP000248482 UP000286642 UP000001593 UP000000715 UP000008912 UP000245320 UP000189704 UP000092124 UP000248483 UP000189706
UP000027135 UP000235965 UP000009046 UP000007819 UP000002358 UP000215335 UP000242457 UP000005203 UP000036403 UP000053825 UP000078540 UP000005205 UP000078542 UP000075809 UP000000311 UP000053097 UP000279307 UP000007755 UP000076502 UP000008237 UP000053105 UP000085678 UP000242188 UP000005408 UP000030746 UP000245119 UP000276133 UP000008854 UP000050788 UP000275846 UP000053454 UP000014760 UP000050789 UP000095280 UP000215902 UP000015101 UP000050795 UP000050740 UP000008672 UP000008227 UP000011712 UP000252040 UP000029965 UP000248482 UP000286642 UP000001593 UP000000715 UP000008912 UP000245320 UP000189704 UP000092124 UP000248483 UP000189706
Pfam
PF10629 DUF2475
Interpro
IPR018902
UPF0573/UPF0605
ProteinModelPortal
A0A2A4JY48
A0A194Q9N8
A0A212EQ83
A0A194QPR0
A0A2H1WZ27
A0A0L7L1A8
+ More
A0A1Y1LJW9 A0A1Y1LH76 A0A139WLN5 A0A067RIT7 A0A1B6L935 A0A1B6M8P7 A0A1B6FUA9 A0A1Y1LIQ8 A0A1Y1LKX4 A0A1Y1LF05 A0A1Y1LH93 A0A2J7RSD9 A0A1B6IB31 E0V9S7 J9LV09 A0A2S2PYG5 A0A1Y1LF43 K7JQ24 A0A232FI72 A0A2A3ES11 A0A088AP87 A0A0J7LAD2 A0A0L7R0H4 A0A195B990 A0A158NL07 A0A195CNS6 A0A151X443 E2ALC4 A0A026WMJ2 F4WB69 A0A154PP91 E2C2X1 A0A0N0U5M9 A0A1S3IG61 A0A210R5D7 K1PU44 A0A0B6YVV5 V4BUX1 C1LFI5 A0A2T7NL09 A0A3M7SU91 A0A075A3B0 A0A177B0G0 H2KV50 Q86DY4 A0A3Q0KKG8 A0A183SZ81 A0A0X3PDQ0 A0A0L8H118 R7UKN5 G4LVN1 B3GUX2 A0A3P7LFY0 A0A2H1C4H5 A0A1S8WLK8 A0A183KFZ5 A0A1I8FW21 A0A1I8FZ71 T1F907 A0A094ZT67 A0A183VVR9 A0A183APF9 M3XJG1 F1SG06 M3WZI7 A0A341CVQ6 A0A0D9RD84 A0A2Y9J762 A0A3Q7X6D7 A0A3Q7XHU8 A7RIM2 A0A3Q7WB28 M3Y1F1 G1LS70 D2HAK2 A0A341CY54 A0A2U4CG61 A0A3Q0E8Y2 A0A1A6HZ27 A0A2Y9QKB1 A0A1U7R146 A0A212EMF4
A0A1Y1LJW9 A0A1Y1LH76 A0A139WLN5 A0A067RIT7 A0A1B6L935 A0A1B6M8P7 A0A1B6FUA9 A0A1Y1LIQ8 A0A1Y1LKX4 A0A1Y1LF05 A0A1Y1LH93 A0A2J7RSD9 A0A1B6IB31 E0V9S7 J9LV09 A0A2S2PYG5 A0A1Y1LF43 K7JQ24 A0A232FI72 A0A2A3ES11 A0A088AP87 A0A0J7LAD2 A0A0L7R0H4 A0A195B990 A0A158NL07 A0A195CNS6 A0A151X443 E2ALC4 A0A026WMJ2 F4WB69 A0A154PP91 E2C2X1 A0A0N0U5M9 A0A1S3IG61 A0A210R5D7 K1PU44 A0A0B6YVV5 V4BUX1 C1LFI5 A0A2T7NL09 A0A3M7SU91 A0A075A3B0 A0A177B0G0 H2KV50 Q86DY4 A0A3Q0KKG8 A0A183SZ81 A0A0X3PDQ0 A0A0L8H118 R7UKN5 G4LVN1 B3GUX2 A0A3P7LFY0 A0A2H1C4H5 A0A1S8WLK8 A0A183KFZ5 A0A1I8FW21 A0A1I8FZ71 T1F907 A0A094ZT67 A0A183VVR9 A0A183APF9 M3XJG1 F1SG06 M3WZI7 A0A341CVQ6 A0A0D9RD84 A0A2Y9J762 A0A3Q7X6D7 A0A3Q7XHU8 A7RIM2 A0A3Q7WB28 M3Y1F1 G1LS70 D2HAK2 A0A341CY54 A0A2U4CG61 A0A3Q0E8Y2 A0A1A6HZ27 A0A2Y9QKB1 A0A1U7R146 A0A212EMF4
Ontologies
GO
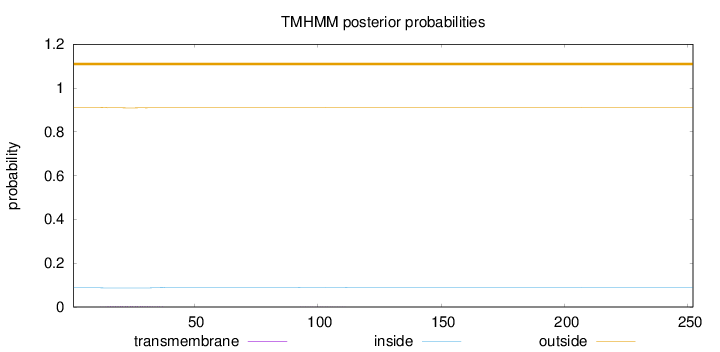
Topology
Length:
252
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07536
Exp number, first 60 AAs:
0.07018
Total prob of N-in:
0.08790
outside
1 - 252
Population Genetic Test Statistics
Pi
23.094541
Theta
20.341455
Tajima's D
0.135989
CLR
1.110671
CSRT
0.410879456027199
Interpretation
Uncertain
