Gene
KWMTBOMO06796
Annotation
PREDICTED:_peptide_transporter_family_1_[Bombyx_mori]
Full name
Solute carrier family 15 member 1
+ More
Peptide transporter family 1
Peptide transporter family 1
Alternative Name
Intestinal H(+)/peptide cotransporter
Oligopeptide transporter, small intestine isoform
Peptide transporter 1
Proton-coupled dipeptide cotransporter
Oligopeptide transporter 1
Protein YIN
Oligopeptide transporter, small intestine isoform
Peptide transporter 1
Proton-coupled dipeptide cotransporter
Oligopeptide transporter 1
Protein YIN
Location in the cell
PlasmaMembrane Reliability : 2.753
Sequence
CDS
ATGACTGAGGAAAAATCGTTTGGCACCAGTAATGGAGTGAATAAGAGTTCGGGTCGTTTTCCCGGAATTGTTATTATTATCATTCTGGCCGAATTTTGTGAACGATTCTCTTATTCGGGAATGAGAGCGTTTTTAACATTATACTTAAGAAGTAAGCTAGGATATACAGACGATGGTGCGACTGAAACATATCACGTTTTTAGTACATTAGTATATGTTTTTCCGATTATTGGAGGCATATTAGCTGATAATTACCTTGGAAAATTCAGGTGA
Protein
MTEEKSFGTSNGVNKSSGRFPGIVIIIILAEFCERFSYSGMRAFLTLYLRSKLGYTDDGATETYHVFSTLVYVFPIIGGILADNYLGKFR
Summary
Description
Important role in absorption of dietary peptides. High-affinity transporter of alanylalanine. Dipeptide transport activity is proton dependent.
Biophysicochemical Properties
48.8 uM for alanylalanine
Similarity
Belongs to the PTR2/POT transporter (TC 2.A.17) family.
Keywords
Alternative promoter usage
Complete proteome
Glycoprotein
Membrane
Peptide transport
Protein transport
Reference proteome
Symport
Transmembrane
Transmembrane helix
Transport
Alternative splicing
Feature
chain Solute carrier family 15 member 1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A3S2LIB9
A0A0L7LC32
A0A212FBF6
A0A194QVB4
A0A194QAU8
A0A1J1HW66
+ More
A0A0A1XQB7 Q16YY6 H9ITB6 F1N3X5 A0A194QN08 A0A1B6IYV0 A0A212EGH0 S4P9F4 W8ANZ8 A0A336MER3 A0A1B0G016 A0A0C9RZK3 A0A2P8YIJ6 A0A0C9QQB9 A0A0A1WEF5 A0A1W5BEF6 B4KF56 A0A1B0DQT0 A0A0K8W9N9 A0A1S4G7L9 A0A0K8U2K2 B4L740 B4MB13 A0A1B6JRC5 A0A0Q9XTK6 A0A3Q2YJG8 A0A1B6J332 A0A034WNV7 A0A1I8Q7P1 X2JAE6 T1DG97 P51574 F1LMZ1 A0A3B0K404 A0A3B0JWR6 T1E1E0 A0A1W2W1N6 A0A2J7R2B1 A0A1Q3EVZ4 A0A1L8E292 F7BLJ0 A0A0R3P597 A0A3B0JRR8 A0A158NT08 B4H2E7 A0A151X6Y5 F4WY69 Q14BA3 A0A182GHS3 A0A151I3X9 A0A2J7R2B4 T1PL08 A0A1I8M995 B0W557 A0A1Q3EVN6 A0A1I8M9A0 A0A3B0J7S6 P91679 B4R478 P91679-3 P91679-2 B4I146 A0A0R3P6G6 A0A182FVA2 A0A3Q3S526 A0A1B6MGR5 B4JX99 A0A0M4F8C2 A0A1A9YF31 A0A0C9PUC0 A0A1B6KAM5 A0A1A9Z201 A0A3R7PF49 Q29GY1 A0A067RAG2 A0A182YC53 B4NDF2 A0A0P4W6I5 A0A3P8Z868 A0A0Q9WUA5 A0A3Q3WB15 A0A182T3W5 A0A0M9A6L9 H2Y3X2 A0A182X015 A0A182VMC2 A0A182KT24 A0A182HPR3 A0A3B5BI36 A0A1B6GZH0 B3MQY6 Q7PZY8 H2Y3X3
A0A0A1XQB7 Q16YY6 H9ITB6 F1N3X5 A0A194QN08 A0A1B6IYV0 A0A212EGH0 S4P9F4 W8ANZ8 A0A336MER3 A0A1B0G016 A0A0C9RZK3 A0A2P8YIJ6 A0A0C9QQB9 A0A0A1WEF5 A0A1W5BEF6 B4KF56 A0A1B0DQT0 A0A0K8W9N9 A0A1S4G7L9 A0A0K8U2K2 B4L740 B4MB13 A0A1B6JRC5 A0A0Q9XTK6 A0A3Q2YJG8 A0A1B6J332 A0A034WNV7 A0A1I8Q7P1 X2JAE6 T1DG97 P51574 F1LMZ1 A0A3B0K404 A0A3B0JWR6 T1E1E0 A0A1W2W1N6 A0A2J7R2B1 A0A1Q3EVZ4 A0A1L8E292 F7BLJ0 A0A0R3P597 A0A3B0JRR8 A0A158NT08 B4H2E7 A0A151X6Y5 F4WY69 Q14BA3 A0A182GHS3 A0A151I3X9 A0A2J7R2B4 T1PL08 A0A1I8M995 B0W557 A0A1Q3EVN6 A0A1I8M9A0 A0A3B0J7S6 P91679 B4R478 P91679-3 P91679-2 B4I146 A0A0R3P6G6 A0A182FVA2 A0A3Q3S526 A0A1B6MGR5 B4JX99 A0A0M4F8C2 A0A1A9YF31 A0A0C9PUC0 A0A1B6KAM5 A0A1A9Z201 A0A3R7PF49 Q29GY1 A0A067RAG2 A0A182YC53 B4NDF2 A0A0P4W6I5 A0A3P8Z868 A0A0Q9WUA5 A0A3Q3WB15 A0A182T3W5 A0A0M9A6L9 H2Y3X2 A0A182X015 A0A182VMC2 A0A182KT24 A0A182HPR3 A0A3B5BI36 A0A1B6GZH0 B3MQY6 Q7PZY8 H2Y3X3
Pubmed
26227816
22118469
26354079
25830018
17510324
19121390
+ More
19393038 23622113 24495485 29403074 17994087 18057021 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24330624 8605246 8531138 15684415 19103603 15057822 12481130 15114417 15632085 21347285 21719571 15489334 26483478 25315136 9038337 9730971 10731137 12537569 24845553 25244985 25069045 20966253 12364791
19393038 23622113 24495485 29403074 17994087 18057021 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24330624 8605246 8531138 15684415 19103603 15057822 12481130 15114417 15632085 21347285 21719571 15489334 26483478 25315136 9038337 9730971 10731137 12537569 24845553 25244985 25069045 20966253 12364791
EMBL
RSAL01000107
RVE47292.1
JTDY01001763
KOB72959.1
AGBW02009331
OWR51072.1
+ More
KQ461185 KPJ07496.1 KQ459252 KPJ02120.1 CVRI01000025 CRK92335.1 GBXI01001222 JAD13070.1 CH477506 EAT39840.1 BABH01007227 KQ461196 KPJ06360.1 GECU01015578 JAS92128.1 AGBW02015069 OWR40591.1 GAIX01005511 JAA87049.1 GAMC01020327 GAMC01020325 JAB86228.1 UFQT01001081 SSX28795.1 CCAG010002596 GBYB01013556 JAG83323.1 PYGN01000569 PSN44085.1 GBYB01002827 JAG72594.1 GBXI01017457 JAC96834.1 CH933807 EDW13039.1 AJVK01019354 AJVK01019355 GDHF01004575 JAI47739.1 GDHF01031581 JAI20733.1 CH933812 EDW06186.1 CH940655 EDW66422.2 GECU01005989 JAT01718.1 KRG07171.1 KRG07172.1 GECU01014134 JAS93572.1 GAKP01003077 JAC55875.1 AE014298 AHN59315.1 GALA01001761 JAA93091.1 D50664 D50306 AY860424 AABR07019460 AABR07019461 OUUW01000003 SPP78178.1 SPP78179.1 GALA01001764 JAA93088.1 NEVH01007841 PNF34977.1 GFDL01015667 JAV19378.1 GFDF01001256 JAV12828.1 CH379064 KRT06725.1 SPP78180.1 ADTU01025466 ADTU01025467 ADTU01025468 ADTU01025469 CH479204 EDW30514.1 KQ982476 KYQ56058.1 GL888439 EGI60826.1 BC116248 BC116249 AAI16250.1 JXUM01064548 JXUM01064549 JXUM01064550 KQ562303 KXJ76196.1 KQ976472 KYM83981.1 PNF34978.1 KA648800 AFP63429.1 DS231841 EDS34804.1 GFDL01015675 JAV19370.1 SPP78177.1 U85982 U97114 AL031130 BT031266 BT031310 AY058467 CM000366 EDX17020.1 CH480819 EDW53227.1 KRT06726.1 GEBQ01004875 JAT35102.1 CH916376 EDV95375.1 CP012528 ALC48232.1 GBYB01004958 JAG74725.1 GEBQ01031478 JAT08499.1 QCYY01003298 ROT64216.1 EAL31978.1 KK852588 KDR20782.1 CH964239 EDW82858.1 GDRN01079346 JAI62419.1 KRF99675.1 KQ435724 KOX78297.1 APCN01003267 GECZ01001950 JAS67819.1 CH902622 EDV34191.2 AAAB01008986 EAA00193.4
KQ461185 KPJ07496.1 KQ459252 KPJ02120.1 CVRI01000025 CRK92335.1 GBXI01001222 JAD13070.1 CH477506 EAT39840.1 BABH01007227 KQ461196 KPJ06360.1 GECU01015578 JAS92128.1 AGBW02015069 OWR40591.1 GAIX01005511 JAA87049.1 GAMC01020327 GAMC01020325 JAB86228.1 UFQT01001081 SSX28795.1 CCAG010002596 GBYB01013556 JAG83323.1 PYGN01000569 PSN44085.1 GBYB01002827 JAG72594.1 GBXI01017457 JAC96834.1 CH933807 EDW13039.1 AJVK01019354 AJVK01019355 GDHF01004575 JAI47739.1 GDHF01031581 JAI20733.1 CH933812 EDW06186.1 CH940655 EDW66422.2 GECU01005989 JAT01718.1 KRG07171.1 KRG07172.1 GECU01014134 JAS93572.1 GAKP01003077 JAC55875.1 AE014298 AHN59315.1 GALA01001761 JAA93091.1 D50664 D50306 AY860424 AABR07019460 AABR07019461 OUUW01000003 SPP78178.1 SPP78179.1 GALA01001764 JAA93088.1 NEVH01007841 PNF34977.1 GFDL01015667 JAV19378.1 GFDF01001256 JAV12828.1 CH379064 KRT06725.1 SPP78180.1 ADTU01025466 ADTU01025467 ADTU01025468 ADTU01025469 CH479204 EDW30514.1 KQ982476 KYQ56058.1 GL888439 EGI60826.1 BC116248 BC116249 AAI16250.1 JXUM01064548 JXUM01064549 JXUM01064550 KQ562303 KXJ76196.1 KQ976472 KYM83981.1 PNF34978.1 KA648800 AFP63429.1 DS231841 EDS34804.1 GFDL01015675 JAV19370.1 SPP78177.1 U85982 U97114 AL031130 BT031266 BT031310 AY058467 CM000366 EDX17020.1 CH480819 EDW53227.1 KRT06726.1 GEBQ01004875 JAT35102.1 CH916376 EDV95375.1 CP012528 ALC48232.1 GBYB01004958 JAG74725.1 GEBQ01031478 JAT08499.1 QCYY01003298 ROT64216.1 EAL31978.1 KK852588 KDR20782.1 CH964239 EDW82858.1 GDRN01079346 JAI62419.1 KRF99675.1 KQ435724 KOX78297.1 APCN01003267 GECZ01001950 JAS67819.1 CH902622 EDV34191.2 AAAB01008986 EAA00193.4
Proteomes
UP000283053
UP000037510
UP000007151
UP000053240
UP000053268
UP000183832
+ More
UP000008820 UP000005204 UP000009136 UP000092444 UP000245037 UP000009192 UP000092462 UP000008792 UP000264820 UP000095300 UP000000803 UP000002494 UP000268350 UP000235965 UP000008144 UP000001819 UP000005205 UP000008744 UP000075809 UP000007755 UP000069940 UP000249989 UP000078540 UP000095301 UP000002320 UP000000304 UP000001292 UP000069272 UP000261640 UP000001070 UP000092553 UP000092443 UP000092445 UP000283509 UP000027135 UP000076408 UP000007798 UP000265140 UP000261620 UP000075901 UP000053105 UP000007875 UP000076407 UP000075903 UP000075882 UP000075840 UP000261400 UP000007801 UP000007062
UP000008820 UP000005204 UP000009136 UP000092444 UP000245037 UP000009192 UP000092462 UP000008792 UP000264820 UP000095300 UP000000803 UP000002494 UP000268350 UP000235965 UP000008144 UP000001819 UP000005205 UP000008744 UP000075809 UP000007755 UP000069940 UP000249989 UP000078540 UP000095301 UP000002320 UP000000304 UP000001292 UP000069272 UP000261640 UP000001070 UP000092553 UP000092443 UP000092445 UP000283509 UP000027135 UP000076408 UP000007798 UP000265140 UP000261620 UP000075901 UP000053105 UP000007875 UP000076407 UP000075903 UP000075882 UP000075840 UP000261400 UP000007801 UP000007062
Pfam
PF00854 PTR2
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A3S2LIB9
A0A0L7LC32
A0A212FBF6
A0A194QVB4
A0A194QAU8
A0A1J1HW66
+ More
A0A0A1XQB7 Q16YY6 H9ITB6 F1N3X5 A0A194QN08 A0A1B6IYV0 A0A212EGH0 S4P9F4 W8ANZ8 A0A336MER3 A0A1B0G016 A0A0C9RZK3 A0A2P8YIJ6 A0A0C9QQB9 A0A0A1WEF5 A0A1W5BEF6 B4KF56 A0A1B0DQT0 A0A0K8W9N9 A0A1S4G7L9 A0A0K8U2K2 B4L740 B4MB13 A0A1B6JRC5 A0A0Q9XTK6 A0A3Q2YJG8 A0A1B6J332 A0A034WNV7 A0A1I8Q7P1 X2JAE6 T1DG97 P51574 F1LMZ1 A0A3B0K404 A0A3B0JWR6 T1E1E0 A0A1W2W1N6 A0A2J7R2B1 A0A1Q3EVZ4 A0A1L8E292 F7BLJ0 A0A0R3P597 A0A3B0JRR8 A0A158NT08 B4H2E7 A0A151X6Y5 F4WY69 Q14BA3 A0A182GHS3 A0A151I3X9 A0A2J7R2B4 T1PL08 A0A1I8M995 B0W557 A0A1Q3EVN6 A0A1I8M9A0 A0A3B0J7S6 P91679 B4R478 P91679-3 P91679-2 B4I146 A0A0R3P6G6 A0A182FVA2 A0A3Q3S526 A0A1B6MGR5 B4JX99 A0A0M4F8C2 A0A1A9YF31 A0A0C9PUC0 A0A1B6KAM5 A0A1A9Z201 A0A3R7PF49 Q29GY1 A0A067RAG2 A0A182YC53 B4NDF2 A0A0P4W6I5 A0A3P8Z868 A0A0Q9WUA5 A0A3Q3WB15 A0A182T3W5 A0A0M9A6L9 H2Y3X2 A0A182X015 A0A182VMC2 A0A182KT24 A0A182HPR3 A0A3B5BI36 A0A1B6GZH0 B3MQY6 Q7PZY8 H2Y3X3
A0A0A1XQB7 Q16YY6 H9ITB6 F1N3X5 A0A194QN08 A0A1B6IYV0 A0A212EGH0 S4P9F4 W8ANZ8 A0A336MER3 A0A1B0G016 A0A0C9RZK3 A0A2P8YIJ6 A0A0C9QQB9 A0A0A1WEF5 A0A1W5BEF6 B4KF56 A0A1B0DQT0 A0A0K8W9N9 A0A1S4G7L9 A0A0K8U2K2 B4L740 B4MB13 A0A1B6JRC5 A0A0Q9XTK6 A0A3Q2YJG8 A0A1B6J332 A0A034WNV7 A0A1I8Q7P1 X2JAE6 T1DG97 P51574 F1LMZ1 A0A3B0K404 A0A3B0JWR6 T1E1E0 A0A1W2W1N6 A0A2J7R2B1 A0A1Q3EVZ4 A0A1L8E292 F7BLJ0 A0A0R3P597 A0A3B0JRR8 A0A158NT08 B4H2E7 A0A151X6Y5 F4WY69 Q14BA3 A0A182GHS3 A0A151I3X9 A0A2J7R2B4 T1PL08 A0A1I8M995 B0W557 A0A1Q3EVN6 A0A1I8M9A0 A0A3B0J7S6 P91679 B4R478 P91679-3 P91679-2 B4I146 A0A0R3P6G6 A0A182FVA2 A0A3Q3S526 A0A1B6MGR5 B4JX99 A0A0M4F8C2 A0A1A9YF31 A0A0C9PUC0 A0A1B6KAM5 A0A1A9Z201 A0A3R7PF49 Q29GY1 A0A067RAG2 A0A182YC53 B4NDF2 A0A0P4W6I5 A0A3P8Z868 A0A0Q9WUA5 A0A3Q3WB15 A0A182T3W5 A0A0M9A6L9 H2Y3X2 A0A182X015 A0A182VMC2 A0A182KT24 A0A182HPR3 A0A3B5BI36 A0A1B6GZH0 B3MQY6 Q7PZY8 H2Y3X3
PDB
4Q65
E-value=5.45641e-10,
Score=147
Ontologies
GO
PANTHER
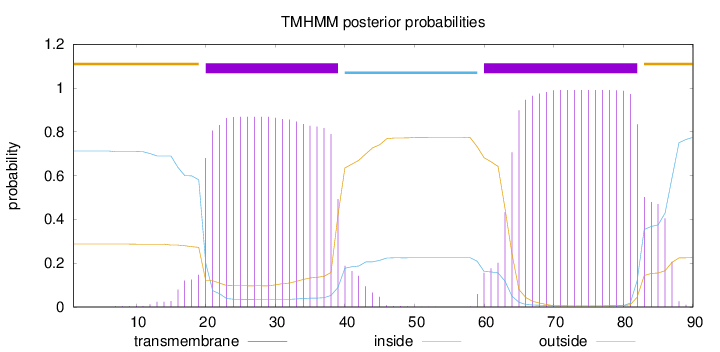
Topology
Subcellular location
Membrane
Length:
90
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
38.98794
Exp number, first 60 AAs:
17.91384
Total prob of N-in:
0.71269
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 39
inside
40 - 59
TMhelix
60 - 82
outside
83 - 90
Population Genetic Test Statistics
Pi
25.106526
Theta
24.831781
Tajima's D
0.089788
CLR
0.310452
CSRT
0.400029998500075
Interpretation
Uncertain
