Gene
KWMTBOMO06792
Pre Gene Modal
BGIBMGA012041
Annotation
neuropeptide_receptor_A11_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.92
Sequence
CDS
ATGTTCGGAAATGATACTCTTCAGGATGTTATGGCCTCAACGATAGCCGCGAAGTATGAACCAGCCATGTCTCTGAACGGGACCACATATGTGGGCGGGGGGGTCCTTATCCTCACTCGGACTTTGACGGGTGAATCAGTTGAGATGATCGACGAACCGAAAACAAACAAAACGATAGACATAATAGACGTGAAATTAGTTCAGGTCGCTTTTTGCATTCTCTACACAGTAATCTTTGTTCTGGGCGTGTTCGGAAATGTCCTTGTTTGCTATGTGGTTTTCCGCAACAAAGCTATGCAAACAGTCACGAACCTTTTCATAACGAACCTGGCTTTATCAGATATATTGCTGTGTGTCTTCGCCGTACCTTTGACGCCAATGTATACGTTTTTGGGACGATGGGTGTTCGGGAGATTGCTCTGTCATCTGATGCCGTATGCACAAGGAACAAGTGTGTACATATCCACGCTGACGTTGACTTCAATAGCCATAGATAGATTTTTCGTGATAATATACCCGTTTCACCCTAGAATGAAACTGAACACTTGTATTTTTATTGTCATTTTCATATGGGTGTTCTCTTTGGTTGTAACATGTCCTTACGGACTATTCATGGGAATACAGACCACCAACAATGAGACGTATTATTGCGAAGAAAGTTGGCCCTCAGACAGATCTAGAAAAATATTTGGCGTATTTACGACAGTTCTTCAGTTTTTAATACCCTTTTTAGTAATAGCTGTATGCTACACGTGTGTGAGTATAAGATTGAATGATCGGGCTCGGTCAAAACCAGGTGCTAAAAATAGCAAAAGGGAAGAAGCGGACAGAGATAGGAAAAGGTGTACTAACAGAATGTTAATTTCAATGGTCGCGATATTTGGAATCTCGTGGCTTCCTTTGAACTTGATAAATATATTCAACGATTTTTATGCTCAGATGACTGAGTGGAACTATTACTTCGTGTCATTCTTCTTGGCCCACTCTATGGCCATGGCTTCGACTTGCTATAATCCATTTCTTTACGCCTGGTTGAATGAAAACTTCAGAAAGGAATTCAAGCAGGTGTTGCCCTTTTTCGAGTCGAATGGAGGCGTAAGGAATAGTTACCATCCAGGACGTGTGCCACCCCACAAAACTAACAAGAACGTTTGTAATGGCAACGAAACTATACAGGAAACTTTACTGGCGTCCTCGTTCAATAGGGGACCTTCAATCAAACAGAGATTCGAAGGGAACGGGAAGAAAGATAATGGAATCGAAGTTGAAAACATTTTGTTGGAGGACAAAACAATCTCAGCTACTTTTCATACAAAGACCGAAAACGTCAATTTACAACTGATCGATGAGGAGTCACATTTCAGCGATCACAGAGACACAAAGTCACCGATATAA
Protein
MFGNDTLQDVMASTIAAKYEPAMSLNGTTYVGGGVLILTRTLTGESVEMIDEPKTNKTIDIIDVKLVQVAFCILYTVIFVLGVFGNVLVCYVVFRNKAMQTVTNLFITNLALSDILLCVFAVPLTPMYTFLGRWVFGRLLCHLMPYAQGTSVYISTLTLTSIAIDRFFVIIYPFHPRMKLNTCIFIVIFIWVFSLVVTCPYGLFMGIQTTNNETYYCEESWPSDRSRKIFGVFTTVLQFLIPFLVIAVCYTCVSIRLNDRARSKPGAKNSKREEADRDRKRCTNRMLISMVAIFGISWLPLNLINIFNDFYAQMTEWNYYFVSFFLAHSMAMASTCYNPFLYAWLNENFRKEFKQVLPFFESNGGVRNSYHPGRVPPHKTNKNVCNGNETIQETLLASSFNRGPSIKQRFEGNGKKDNGIEVENILLEDKTISATFHTKTENVNLQLIDEESHFSDHRDTKSPI
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
H9JR81
B3XXM4
A0A2A4K1Y2
A0A2H1W6Q0
A0A0S1YDL2
A0A0L7KXV9
+ More
A0A1E1WNB6 A0A194QPN4 A0A194Q9C5 A0A212ELB7 A0A0S1YDT6 A0A2A4JAM4 H9JRF4 B3XXM3 A0A0L7KGG5 A0A2H1WDX8 N6U615 A0A0L7L7M3 A0A182G4E1 Q16IY5 Q170F7 U5N1G3 U5N412 U5N0U1 A0A182MUV2 W5J9H0 A0A084WIC4 A0A182WF01 A0A182RFF4 B0X0M0 A0A2R4A6V7 A0A336MPB5 A0A087ZV05 A0A182J609 A0A2P8YC91 Q7Q0B9 A0A182FBA6 A0A182WZR5 A0A182K1A6 A0A182QHF8 A0A182KSD4 A0A182VA59 A0SIF2 A0A182HQB0 A0A1B0CPY1 A0A182TJJ9 A0A023F668 A0A182MYV0 A0A182SAA9 A0A0J7NSP3 A0A182PKC0 A0A182Y1J7 A0A2J7Q7Q7 A0A2A3E7C9 E2C1I7 H9JRF2 A0A151XBE0 B3XXM0 A0A195B6I0 A0A158NIG7 A0A195DF11 F4WCK3 A0A195ESR8 A0A1L8DJJ0 A0A139WLV7 A0A1L8DJY6 A0A1B6FAU8 A0A1B6JLJ1 E2A6J5 A0A2A4JY03 A0A1W4XQD5 A0A026WW28 A0A2H1WMM8 A0A1B6LCN2 A0A1B6DKU7 A0A0S1YDF2 A0A0L7R0H5 A0A195C3U0 A0A0C9RH27 A0A232FLS4 A0A1I8NNA2 A0A154PRA0 A0A226E6K8 A0A1D2N5F0 K7J333 A0A0M3QX42 U3U8Y2 A0A0K8TJW6 A0A0L7KYZ8 A0A1A9VHR1 B4LFY2 B4IYT2 B3M3J6 B4KV51 B4MKH9 A0A0J9RYF2 A0A1A9W8G3 A0A194Q9E0 E0VVA3 B4QRF6
A0A1E1WNB6 A0A194QPN4 A0A194Q9C5 A0A212ELB7 A0A0S1YDT6 A0A2A4JAM4 H9JRF4 B3XXM3 A0A0L7KGG5 A0A2H1WDX8 N6U615 A0A0L7L7M3 A0A182G4E1 Q16IY5 Q170F7 U5N1G3 U5N412 U5N0U1 A0A182MUV2 W5J9H0 A0A084WIC4 A0A182WF01 A0A182RFF4 B0X0M0 A0A2R4A6V7 A0A336MPB5 A0A087ZV05 A0A182J609 A0A2P8YC91 Q7Q0B9 A0A182FBA6 A0A182WZR5 A0A182K1A6 A0A182QHF8 A0A182KSD4 A0A182VA59 A0SIF2 A0A182HQB0 A0A1B0CPY1 A0A182TJJ9 A0A023F668 A0A182MYV0 A0A182SAA9 A0A0J7NSP3 A0A182PKC0 A0A182Y1J7 A0A2J7Q7Q7 A0A2A3E7C9 E2C1I7 H9JRF2 A0A151XBE0 B3XXM0 A0A195B6I0 A0A158NIG7 A0A195DF11 F4WCK3 A0A195ESR8 A0A1L8DJJ0 A0A139WLV7 A0A1L8DJY6 A0A1B6FAU8 A0A1B6JLJ1 E2A6J5 A0A2A4JY03 A0A1W4XQD5 A0A026WW28 A0A2H1WMM8 A0A1B6LCN2 A0A1B6DKU7 A0A0S1YDF2 A0A0L7R0H5 A0A195C3U0 A0A0C9RH27 A0A232FLS4 A0A1I8NNA2 A0A154PRA0 A0A226E6K8 A0A1D2N5F0 K7J333 A0A0M3QX42 U3U8Y2 A0A0K8TJW6 A0A0L7KYZ8 A0A1A9VHR1 B4LFY2 B4IYT2 B3M3J6 B4KV51 B4MKH9 A0A0J9RYF2 A0A1A9W8G3 A0A194Q9E0 E0VVA3 B4QRF6
Pubmed
19121390
18725956
26227816
26354079
22118469
23537049
+ More
26483478 17510324 24130914 29550617 20920257 23761445 24438588 29403074 12364791 20966253 17140700 25474469 25244985 20798317 21347285 21719571 18362917 19820115 24508170 30249741 28648823 27289101 20075255 23932938 26823975 17994087 22936249 20566863
26483478 17510324 24130914 29550617 20920257 23761445 24438588 29403074 12364791 20966253 17140700 25474469 25244985 20798317 21347285 21719571 18362917 19820115 24508170 30249741 28648823 27289101 20075255 23932938 26823975 17994087 22936249 20566863
EMBL
BABH01016327
KT224452
ALP48446.1
AB330432
BAG68410.1
NWSH01000283
+ More
PCG77773.1 ODYU01006678 SOQ48738.1 KT031009 ALM88307.1 JTDY01004673 KOB67886.1 GDQN01002554 JAT88500.1 KQ461185 KPJ07498.1 KQ459252 KPJ02117.1 AGBW02014093 OWR42269.1 KT031008 ALM88306.1 NWSH01002352 PCG68470.1 BABH01016333 AB330431 BAG68409.1 JTDY01009857 KOB62422.1 ODYU01007985 SOQ51227.1 APGK01041383 KB740993 ENN76071.1 JTDY01002496 KOB71291.1 JXUM01042212 KQ561315 KXJ79017.1 CH478043 EAT34232.2 CH477474 EAT40343.2 KC439530 AGX84998.1 KC439531 AGX84999.1 KC439528 KC439529 MG766139 AGX84996.1 AGX84997.1 AVR59278.1 AXCM01000343 ADMH02001995 ETN60053.1 ATLV01023928 KE525347 KFB49968.1 DS232241 EDS38181.1 MG766141 AVR59280.1 UFQS01001340 UFQS01001735 UFQT01001340 UFQT01001735 SSX10421.1 SSX12089.1 SSX30107.1 PYGN01000709 PSN41879.1 AAAB01008986 EAA00213.1 AXCN02001021 DQ437579 MG766140 ABD96049.1 AVR59279.1 APCN01003304 AJWK01022795 AJWK01022796 GBBI01002004 JAC16708.1 LBMM01001981 KMQ95450.1 NEVH01017441 PNF24618.1 KZ288345 PBC27615.1 GL451937 EFN78163.1 BABH01016315 KQ982327 KYQ57696.1 AB330428 BAG68406.1 KQ976582 KYM79865.1 ADTU01001795 ADTU01001796 KQ980905 KYN11488.1 GL888070 EGI68183.1 KQ981979 KYN31278.1 GFDF01007468 JAV06616.1 KQ971319 KYB28815.1 GFDF01007397 JAV06687.1 GECZ01022420 JAS47349.1 GECU01007570 JAT00137.1 GL437132 EFN70952.1 NWSH01000397 PCG76689.1 KK107078 QOIP01000011 EZA60290.1 RLU17192.1 ODYU01009710 SOQ54331.1 GEBQ01018538 JAT21439.1 GEDC01010982 JAS26316.1 KT031005 ALM88303.1 KQ414670 KOC64326.1 KQ978292 KYM95529.1 GBYB01012507 JAG82274.1 NNAY01000042 OXU31612.1 KQ435007 KZC13640.1 LNIX01000006 OXA53243.1 LJIJ01000202 ODN00510.1 CP012525 ALC45209.1 AB817290 BAO01057.1 GBRD01000065 GDHC01021596 JAG65756.1 JAP97032.1 JTDY01004332 KOB68286.1 CH940647 EDW70381.1 CH916366 EDV96619.1 CH902618 EDV39257.1 CH933809 EDW19391.1 CH963847 EDW72685.2 CM002912 KMZ00682.1 KPJ02118.1 DS235806 EEB17309.1 CM000363 EDX11169.1
PCG77773.1 ODYU01006678 SOQ48738.1 KT031009 ALM88307.1 JTDY01004673 KOB67886.1 GDQN01002554 JAT88500.1 KQ461185 KPJ07498.1 KQ459252 KPJ02117.1 AGBW02014093 OWR42269.1 KT031008 ALM88306.1 NWSH01002352 PCG68470.1 BABH01016333 AB330431 BAG68409.1 JTDY01009857 KOB62422.1 ODYU01007985 SOQ51227.1 APGK01041383 KB740993 ENN76071.1 JTDY01002496 KOB71291.1 JXUM01042212 KQ561315 KXJ79017.1 CH478043 EAT34232.2 CH477474 EAT40343.2 KC439530 AGX84998.1 KC439531 AGX84999.1 KC439528 KC439529 MG766139 AGX84996.1 AGX84997.1 AVR59278.1 AXCM01000343 ADMH02001995 ETN60053.1 ATLV01023928 KE525347 KFB49968.1 DS232241 EDS38181.1 MG766141 AVR59280.1 UFQS01001340 UFQS01001735 UFQT01001340 UFQT01001735 SSX10421.1 SSX12089.1 SSX30107.1 PYGN01000709 PSN41879.1 AAAB01008986 EAA00213.1 AXCN02001021 DQ437579 MG766140 ABD96049.1 AVR59279.1 APCN01003304 AJWK01022795 AJWK01022796 GBBI01002004 JAC16708.1 LBMM01001981 KMQ95450.1 NEVH01017441 PNF24618.1 KZ288345 PBC27615.1 GL451937 EFN78163.1 BABH01016315 KQ982327 KYQ57696.1 AB330428 BAG68406.1 KQ976582 KYM79865.1 ADTU01001795 ADTU01001796 KQ980905 KYN11488.1 GL888070 EGI68183.1 KQ981979 KYN31278.1 GFDF01007468 JAV06616.1 KQ971319 KYB28815.1 GFDF01007397 JAV06687.1 GECZ01022420 JAS47349.1 GECU01007570 JAT00137.1 GL437132 EFN70952.1 NWSH01000397 PCG76689.1 KK107078 QOIP01000011 EZA60290.1 RLU17192.1 ODYU01009710 SOQ54331.1 GEBQ01018538 JAT21439.1 GEDC01010982 JAS26316.1 KT031005 ALM88303.1 KQ414670 KOC64326.1 KQ978292 KYM95529.1 GBYB01012507 JAG82274.1 NNAY01000042 OXU31612.1 KQ435007 KZC13640.1 LNIX01000006 OXA53243.1 LJIJ01000202 ODN00510.1 CP012525 ALC45209.1 AB817290 BAO01057.1 GBRD01000065 GDHC01021596 JAG65756.1 JAP97032.1 JTDY01004332 KOB68286.1 CH940647 EDW70381.1 CH916366 EDV96619.1 CH902618 EDV39257.1 CH933809 EDW19391.1 CH963847 EDW72685.2 CM002912 KMZ00682.1 KPJ02118.1 DS235806 EEB17309.1 CM000363 EDX11169.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000019118 UP000069940 UP000249989 UP000008820 UP000075883 UP000000673 UP000030765 UP000075920 UP000075900 UP000002320 UP000005203 UP000075880 UP000245037 UP000007062 UP000069272 UP000076407 UP000075881 UP000075886 UP000075882 UP000075903 UP000075840 UP000092461 UP000075902 UP000075884 UP000075901 UP000036403 UP000075885 UP000076408 UP000235965 UP000242457 UP000008237 UP000075809 UP000078540 UP000005205 UP000078492 UP000007755 UP000078541 UP000007266 UP000000311 UP000192223 UP000053097 UP000279307 UP000053825 UP000078542 UP000215335 UP000095300 UP000076502 UP000198287 UP000094527 UP000002358 UP000092553 UP000078200 UP000008792 UP000001070 UP000007801 UP000009192 UP000007798 UP000091820 UP000009046 UP000000304
UP000019118 UP000069940 UP000249989 UP000008820 UP000075883 UP000000673 UP000030765 UP000075920 UP000075900 UP000002320 UP000005203 UP000075880 UP000245037 UP000007062 UP000069272 UP000076407 UP000075881 UP000075886 UP000075882 UP000075903 UP000075840 UP000092461 UP000075902 UP000075884 UP000075901 UP000036403 UP000075885 UP000076408 UP000235965 UP000242457 UP000008237 UP000075809 UP000078540 UP000005205 UP000078492 UP000007755 UP000078541 UP000007266 UP000000311 UP000192223 UP000053097 UP000279307 UP000053825 UP000078542 UP000215335 UP000095300 UP000076502 UP000198287 UP000094527 UP000002358 UP000092553 UP000078200 UP000008792 UP000001070 UP000007801 UP000009192 UP000007798 UP000091820 UP000009046 UP000000304
Pfam
PF00001 7tm_1
ProteinModelPortal
H9JR81
B3XXM4
A0A2A4K1Y2
A0A2H1W6Q0
A0A0S1YDL2
A0A0L7KXV9
+ More
A0A1E1WNB6 A0A194QPN4 A0A194Q9C5 A0A212ELB7 A0A0S1YDT6 A0A2A4JAM4 H9JRF4 B3XXM3 A0A0L7KGG5 A0A2H1WDX8 N6U615 A0A0L7L7M3 A0A182G4E1 Q16IY5 Q170F7 U5N1G3 U5N412 U5N0U1 A0A182MUV2 W5J9H0 A0A084WIC4 A0A182WF01 A0A182RFF4 B0X0M0 A0A2R4A6V7 A0A336MPB5 A0A087ZV05 A0A182J609 A0A2P8YC91 Q7Q0B9 A0A182FBA6 A0A182WZR5 A0A182K1A6 A0A182QHF8 A0A182KSD4 A0A182VA59 A0SIF2 A0A182HQB0 A0A1B0CPY1 A0A182TJJ9 A0A023F668 A0A182MYV0 A0A182SAA9 A0A0J7NSP3 A0A182PKC0 A0A182Y1J7 A0A2J7Q7Q7 A0A2A3E7C9 E2C1I7 H9JRF2 A0A151XBE0 B3XXM0 A0A195B6I0 A0A158NIG7 A0A195DF11 F4WCK3 A0A195ESR8 A0A1L8DJJ0 A0A139WLV7 A0A1L8DJY6 A0A1B6FAU8 A0A1B6JLJ1 E2A6J5 A0A2A4JY03 A0A1W4XQD5 A0A026WW28 A0A2H1WMM8 A0A1B6LCN2 A0A1B6DKU7 A0A0S1YDF2 A0A0L7R0H5 A0A195C3U0 A0A0C9RH27 A0A232FLS4 A0A1I8NNA2 A0A154PRA0 A0A226E6K8 A0A1D2N5F0 K7J333 A0A0M3QX42 U3U8Y2 A0A0K8TJW6 A0A0L7KYZ8 A0A1A9VHR1 B4LFY2 B4IYT2 B3M3J6 B4KV51 B4MKH9 A0A0J9RYF2 A0A1A9W8G3 A0A194Q9E0 E0VVA3 B4QRF6
A0A1E1WNB6 A0A194QPN4 A0A194Q9C5 A0A212ELB7 A0A0S1YDT6 A0A2A4JAM4 H9JRF4 B3XXM3 A0A0L7KGG5 A0A2H1WDX8 N6U615 A0A0L7L7M3 A0A182G4E1 Q16IY5 Q170F7 U5N1G3 U5N412 U5N0U1 A0A182MUV2 W5J9H0 A0A084WIC4 A0A182WF01 A0A182RFF4 B0X0M0 A0A2R4A6V7 A0A336MPB5 A0A087ZV05 A0A182J609 A0A2P8YC91 Q7Q0B9 A0A182FBA6 A0A182WZR5 A0A182K1A6 A0A182QHF8 A0A182KSD4 A0A182VA59 A0SIF2 A0A182HQB0 A0A1B0CPY1 A0A182TJJ9 A0A023F668 A0A182MYV0 A0A182SAA9 A0A0J7NSP3 A0A182PKC0 A0A182Y1J7 A0A2J7Q7Q7 A0A2A3E7C9 E2C1I7 H9JRF2 A0A151XBE0 B3XXM0 A0A195B6I0 A0A158NIG7 A0A195DF11 F4WCK3 A0A195ESR8 A0A1L8DJJ0 A0A139WLV7 A0A1L8DJY6 A0A1B6FAU8 A0A1B6JLJ1 E2A6J5 A0A2A4JY03 A0A1W4XQD5 A0A026WW28 A0A2H1WMM8 A0A1B6LCN2 A0A1B6DKU7 A0A0S1YDF2 A0A0L7R0H5 A0A195C3U0 A0A0C9RH27 A0A232FLS4 A0A1I8NNA2 A0A154PRA0 A0A226E6K8 A0A1D2N5F0 K7J333 A0A0M3QX42 U3U8Y2 A0A0K8TJW6 A0A0L7KYZ8 A0A1A9VHR1 B4LFY2 B4IYT2 B3M3J6 B4KV51 B4MKH9 A0A0J9RYF2 A0A1A9W8G3 A0A194Q9E0 E0VVA3 B4QRF6
PDB
5ZBQ
E-value=2.51186e-41,
Score=425
Ontologies
KEGG
GO
Topology
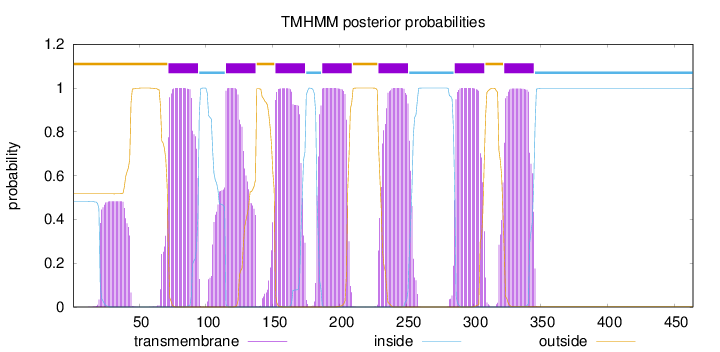
Length:
464
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
167.9391
Exp number, first 60 AAs:
10.55883
Total prob of N-in:
0.48313
POSSIBLE N-term signal
sequence
outside
1 - 71
TMhelix
72 - 94
inside
95 - 114
TMhelix
115 - 137
outside
138 - 151
TMhelix
152 - 174
inside
175 - 186
TMhelix
187 - 209
outside
210 - 228
TMhelix
229 - 251
inside
252 - 285
TMhelix
286 - 308
outside
309 - 322
TMhelix
323 - 345
inside
346 - 464
Population Genetic Test Statistics
Pi
24.794062
Theta
20.175547
Tajima's D
-1.505697
CLR
0.835488
CSRT
0.0589470526473676
Interpretation
Uncertain
