Gene
KWMTBOMO06791
Annotation
PREDICTED:_uncharacterized_protein_K02A2.6-like_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.135
Sequence
CDS
ATGCCGATTGGGAAAGTTGAAAGTTTTGATATGGGATCTCAAAATTGGGACACATACATTCGTCGTGTAAAACAATTCATGGAACTCAACGACATTGAACAGAGACTTCACGTTGCAACGCTCGTAACCTTAGTTGGGTCTGAGTGCTATGACTTAATGTGTGACTTGTGTGCTCCGGACTTACCAGAAGAAAAGGATTTCGACATACTCGTAAAATTAGTCAAAGATCATCTAGAACCTGAAAGATCGGAAATAGCAGAACGTCATATCTTCAGGCAAAGAAAGCAGCAAAATAACGAAAGCATTCGTTCCTACCTACAAAGTTTGAAACATTTGGCTAAAACTTGCAATTTCGGCATCACATTAGAAATTAATCTACGCGATCAATTCGTATCAGGATTATATAGTGAGGAGATGCGATCAAGGCTGTTTGCTGAAAAAGATATAGATTACAAGCGCGCTGTTGAGTTGGCATCAGCACTGGAAGCAGCGGATCGACATGCGATGGCGGCTGGGGGCACTGGGGGCAGCTCACAGGCTGCGTCCGAGAGCGATGGCATGCATCGGGTCGGCGGAGCGCGCGCGGAGGCTCGCTGCGCCTGCAGACGCTGCGGCAAGCTGGGCCATGCGGAGGGTCGGTGTCGGTACAAGCACTACACCTGTGACTCATGTGGCGAGAAGGGACACCTAAAATCAATGTGTCGGAACCGTAAAGGTAACCAAAAGATTGAAAAAAACCGAGGTGGTGGTGAATATTACACAAAATTGGATTTATCTAATGCATTCCAGCAATGCCTCTTAGACGAAGAATCCCAGCCGATGACAGCGATCACGACACACATTGGTACTTTTGTCTATAAGCGCGTTCCGTTTGGAATTAAATGCATTCCGGAAAACTTTCAGAAGATTATGGAAGAGATTCTAAGTGGTCTGCCGTCAACCGTTATTTTTGCGGACGATATATGTGTAACCGGAAAAAACAGTTATATTCTTGGAAAGAATAAAGGTATACCTCTAACCGCCGCGAGTCGTCTGCAACGATTTGCTGTAAAGCTAGCTGCGTATGATTTTGAGATCGAATTTATTAGTTCTACAGATAATTGCCAGGCTGACGCGCTATCACGACTTCCACTGGATACTAATTTTAAGAATAATATTAAGGAGTATAATTACCTAAACTTCGTTCAAGAAAGCTTTCCACTCAGTTTTAAAGAAATTAAAAAGGAAGTTACGAAGGATCCTTTACTAAACAAAATTTACGGTTACGTCATGTTTGGCTGGCCCTCTGTCAGTAAAGAACAAGCAGAAAAACCCTATTTTAACAGAAGAGAATCCATACATATTGATCACGGTTGTCTTGTATGGGGTTACAGAATAATAATTCCAACTTCATTGCGAGAGACTGTGTTACAGGAGTTACATGAGGGACATCCGGGTATTGTTAAGATGAAACAATTAAGCCGGAACTACGTATGGTGGGAGACAGTCGATGCAGACATTGAACGTGTATGTGCTCAGTGCCAGGCCTGTCGATCTCAGCGCGCTGCACCACCATCGGTGCCGCTGCACTCGTGGCCATGGCCGGAAGAGCCGTGGGCGCGCCTTAATCTTGATTTCTTGGGACCGTTTAATAACAAATATTACTTAATTATAGTGGACGCTCATTCAAAATGGATCGAGGTGGAAAAATTGAGCTGTACATCAGCCACGGCAGTAATCGCATGTCTCAGACGATTGTTTGCTCGATTTGGCTTAGCAAAAAAGGTAGTAACTGATAACGGACCACCTTTCTCGTCGATCGAATTTAGAACATATTTAAGTAAAAACGGTATAAAACATTTACCGGTGGCTCCGTATCATCCATCTAGCAATGGTGCAGCGGAAAACGCCGTAAAAACCATAAAAAGAGTGTTGAAAAAGGCTTTAATTGAAAAGGAAGACGACAATACCGCCTTGACAAAATTTCTCTTTATGTACAGAAATTCTGAGCACAGCACTACAGGGCGCGAACCAGCAGTTGCTTTGTTCGGACGTCGCCTGCGTGGAAGACTGGATCTGTTGCGTCCAAATACTAGCTCCATAGTACGAGAAGCACAGCTGGCAAGCGAAAGCTATAATGCAGGCACAGCCGGATATAGGGAGGCGGCAGAAGGAGACTCGGTCCTATTGCAAGATTTCACACAGGGAAAAAAGAAATGGACCACAGGAACAATAAAAAACCGCTCAGGTCCGGTGACATACAGGGTAACAGCAGATGACGGGCGGGTACACAAACGTCACATCGACCAGATTTTAATAAGTAACAATAGTCGAAAATCACGTTATTCTTTGTCCAAATTAAACACTGATTTCGAGACAAAATCAGTAAATGATAGTGTGGGTGAAGTTCGGGAAAGTCCGTGTATGACCGGAGAAAAGGAGCTCAACTCCAGCTATGACACGGGACCGGAAAGTCCACAACCTGTGACTCCTAGTCACTCGCCGGCGCCGAATGTCTTGCAACCATCGCGTCAGCGCCGTAAAGCTGCTTTAACGTGTATTCAGAGAATTAAGGACCAAAATTACTAA
Protein
MPIGKVESFDMGSQNWDTYIRRVKQFMELNDIEQRLHVATLVTLVGSECYDLMCDLCAPDLPEEKDFDILVKLVKDHLEPERSEIAERHIFRQRKQQNNESIRSYLQSLKHLAKTCNFGITLEINLRDQFVSGLYSEEMRSRLFAEKDIDYKRAVELASALEAADRHAMAAGGTGGSSQAASESDGMHRVGGARAEARCACRRCGKLGHAEGRCRYKHYTCDSCGEKGHLKSMCRNRKGNQKIEKNRGGGEYYTKLDLSNAFQQCLLDEESQPMTAITTHIGTFVYKRVPFGIKCIPENFQKIMEEILSGLPSTVIFADDICVTGKNSYILGKNKGIPLTAASRLQRFAVKLAAYDFEIEFISSTDNCQADALSRLPLDTNFKNNIKEYNYLNFVQESFPLSFKEIKKEVTKDPLLNKIYGYVMFGWPSVSKEQAEKPYFNRRESIHIDHGCLVWGYRIIIPTSLRETVLQELHEGHPGIVKMKQLSRNYVWWETVDADIERVCAQCQACRSQRAAPPSVPLHSWPWPEEPWARLNLDFLGPFNNKYYLIIVDAHSKWIEVEKLSCTSATAVIACLRRLFARFGLAKKVVTDNGPPFSSIEFRTYLSKNGIKHLPVAPYHPSSNGAAENAVKTIKRVLKKALIEKEDDNTALTKFLFMYRNSEHSTTGREPAVALFGRRLRGRLDLLRPNTSSIVREAQLASESYNAGTAGYREAAEGDSVLLQDFTQGKKKWTTGTIKNRSGPVTYRVTADDGRVHKRHIDQILISNNSRKSRYSLSKLNTDFETKSVNDSVGEVRESPCMTGEKELNSSYDTGPESPQPVTPSHSPAPNVLQPSRQRRKAALTCIQRIKDQNY
Summary
Uniprot
EMBL
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
PDB
3S3N
E-value=1.31522e-09,
Score=154
Ontologies
KEGG
GO
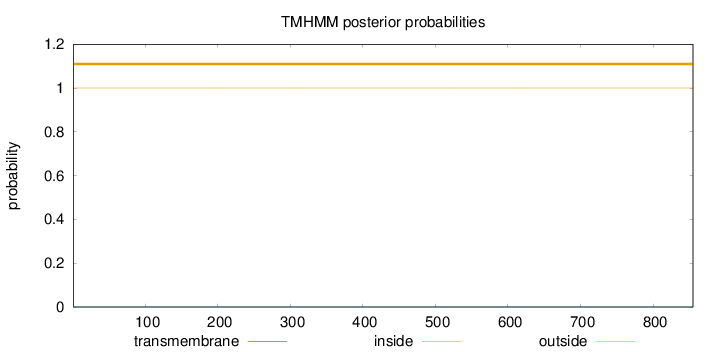
Topology
Length:
855
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00059
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00003
outside
1 - 855
Population Genetic Test Statistics
Pi
10.603958
Theta
21.414367
Tajima's D
-0.856454
CLR
295.078218
CSRT
0.164341782910854
Interpretation
Uncertain
