Gene
KWMTBOMO06787 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012116
Annotation
PREDICTED:_SUMO-activating_enzyme_subunit_2_[Amyelois_transitella]
Full name
SUMO-activating enzyme subunit
Location in the cell
Cytoplasmic Reliability : 2.149 Nuclear Reliability : 1.709
Sequence
CDS
ATGGTCGCGAGGGTCGCTGGCGTTTTCGATGAGAAATTGACAGAAGCTATTGCAAATTCTAAATTGCTGGTAGTGGGCGCCGGCGGCATTGGTTGTGAGATCCTTAAAAATCTCGTGCTAACCGGCTTCCCATATATTGAAATTATCGACCTTGATACTATCGACGTAAGCAACTTGAATCGTCAGTTTCTTTTTCACAAAGAGCATGTTGGGAAATCAAAGGCTCAAGTTGCAAAGGATAGCGCCCTTAGTTTCAACCCTAACGTAACAATCGTAGCTCACCATGACAGTGTAATCAGCAATGACTATGGTGTAAAATACTTCAAACAGTTCAACATTGTGATGAATGCACTGGACAATCGTGTTGCCCGTAACCATGTTAATAGATTGTGTCTTGCTGCTGATGTACCTCTTATAGAGACAGGTACAGCAGGATATGCTGGTCAGGTGGAGCTCATCAAGAAAGGTCTGACTCAATGTTATGAATGCCAGCCTAAAGCACCACAGAAATCATTCCCAGGTTGCACAATTCGTAATACACCTTCAGAACCTATTCACTGCATTGTATGGGCAAAACACCTCTTCAACCAATTGTTTGGAGAAGAAGACCCTGATCAAGATGTGAGTCCTGATACAGCTGACCCTGAGGCTGCAGGTGATGCTGGACAATCAGCGTTGTCTAATGAAAGTACTACTGCCGGTAATGTTGAAAGAAAAAGTACCAGAGCCTGGGCAGCAGAAACTAATTATGAACCAGAAAAGTTATTCACAAAATTATTTGGTGATGATATCCGTTACTTACTATCTATGGAGAATTTGTGGAAGAAACGTCGGCCTCCCACTCCACTGTCATGGGAAAATCTACCTGGAAAAGATGCTCCAAAAATTCAACATTCAGGGTTGCCAGACCAAAGAGTTTGGACAGTATATGAATGTGCACAAGTATTTGCAGCAAGCTGCAAGGCTCTACAGACAGATTTGAAATCGCGTCCGGGTGACGACCATCTTGTATGGGACAAAGATGAAAAAAGTGCCATGGATTTTGTAACTGCCTGTGCTAACATACGCGCACATATCTTTAATATACCACTAAAATCACGATTCGAAATCAAATCAATGGCAGGTAACATCATTCCGGCAATCGCAACGGCCAATGCAATAGTGGCTGGTCTGGCTGTGTTACGTGCACAGTCCATACTGAAAGGAGAAATAGAAAGCTGCACTAGTGTGTACTTGAGACCTAAAGTAAACCACAGGGGACAGCTTTTTGTTCCTGAAAAAACATTAACTCCACCTAATCCTAAGTGCTATGTCTGTCCCCCAAAGCCAGAAGTGGCTCTAGCATGCAACTTGAAAAAACTCACATTGAAAGACCTAGATACGGCTTTCAAAGAGGGCTTAAACATGCAAGCCCCAGACGCCAGTGTAGAAGGCAAAGGTCTTGTTATTTTATCTTCCGAACCAGGAGAAACTGAACACAACAATGACAAAACTCTGGAACAAATCGGCTTGAATGACGGCTGTGCATTACTTGTGGATGATTTCTTACAAAACTATGAAGTCAGGGTCAGACTGCAGCAGGAAGATGAAGAAAATTCTTGGCGGCTGGTAACAGATACCGATGAACCGATGCCGGCGCCTAAAGCAGCAGAAAATTCCGCTAACGGCTCCAATGGCGCTAAGGATCCTGCACCAGGACCTTCCAGATCTCGCCAGGACAGCGACAGTGATATGGAGATAATCGAGGAAGATGACGACGAACCCACAATGAAACCACCTAAGCGCAGGCGTGTCGAAATGTCAGATGAAGTTGTCGAAATATGTTAA
Protein
MVARVAGVFDEKLTEAIANSKLLVVGAGGIGCEILKNLVLTGFPYIEIIDLDTIDVSNLNRQFLFHKEHVGKSKAQVAKDSALSFNPNVTIVAHHDSVISNDYGVKYFKQFNIVMNALDNRVARNHVNRLCLAADVPLIETGTAGYAGQVELIKKGLTQCYECQPKAPQKSFPGCTIRNTPSEPIHCIVWAKHLFNQLFGEEDPDQDVSPDTADPEAAGDAGQSALSNESTTAGNVERKSTRAWAAETNYEPEKLFTKLFGDDIRYLLSMENLWKKRRPPTPLSWENLPGKDAPKIQHSGLPDQRVWTVYECAQVFAASCKALQTDLKSRPGDDHLVWDKDEKSAMDFVTACANIRAHIFNIPLKSRFEIKSMAGNIIPAIATANAIVAGLAVLRAQSILKGEIESCTSVYLRPKVNHRGQLFVPEKTLTPPNPKCYVCPPKPEVALACNLKKLTLKDLDTAFKEGLNMQAPDASVEGKGLVILSSEPGETEHNNDKTLEQIGLNDGCALLVDDFLQNYEVRVRLQQEDEENSWRLVTDTDEPMPAPKAAENSANGSNGAKDPAPGPSRSRQDSDSDMEIIEEDDDEPTMKPPKRRRVEMSDEVVEIC
Summary
Subunit
Heterodimer.
Similarity
Belongs to the ubiquitin-activating E1 family.
Uniprot
H9JRF6
A0A2H1V9X0
A0A194Q9D4
A0A212EK97
A0A3S2LYU5
S4P6Q4
+ More
A0A2A4J3G1 A0A2J7QGR2 A0A067R5B3 A0A088ARE4 A0A2A3EE79 A0A232EM52 A0A0M9A6E1 K7IM19 A0A0L7RIJ3 A0A1B6J1A6 Q16SB9 A0A023EVG3 E9J4H9 A0A182GE15 A0A158NM78 A0A151JWD3 B0WLL0 F4WK04 A0A1Q3F0M9 E0VL42 E2A8Z0 A0A026WHT9 A0A151IRI2 A0A151X2E0 A0A151IE81 A0A0J7L327 A0A195BYV4 A0A182RK53 E2B7T3 A0A0C9QK38 U5EXW5 A0A336K919 A0A182LUB3 A0A182WFG0 A0A1B6F7D1 A0A182YSB3 A0A182JX61 A0A182X5W3 W5JJ08 A0A182SCW5 A0A182VC82 A0A182KN71 A0A182IBJ6 A0A2M4BGD2 A0A2M4BG96 A0A2M4BGA4 A0A182TP29 Q7Q8G9 A0A182P488 A0A2M4AIY1 A0A182ISB0 A0A2M4AJ56 A0A210PH49 A0A182NGP2 A0A3Q0J3U9 A0A084W7N9 A0A182Q1M4 A0A2C9K596 A0A1S3J3I5 A0A2R2MPD2 A0A1S3J4Q2 A0A0B7BGF8 A0A1J1HST2 A0A1B0CYM6 K1Q5G9 A0A0L0C1Y7 A0A1B6DVJ7 V4ATI1 A0A1L8DST4 A0A0T6B393 A0A2T7NHS4 A0A1A9ZX08 A0A2B4RUV6 A0A1W4WFV1 A0A1Y1KF70 A0A1B0G3Y6 A0A1A9XIL0 A0A1B0BCP7 A0A3M6U7E3 A0A0N8DRQ8 A0A1I8PHE9 A0A0P5E456 A0A0P4XPT0 A0A0N8CL88 A0A0P5ZWY1 A0A0P4YU31 A0A0P6E5M0 A0A165A5B7 A0A1I8MXD0 A0A0P5XLD6 A0A1A9XPX7 A0A1A9VN27 A0A1B0G2L0
A0A2A4J3G1 A0A2J7QGR2 A0A067R5B3 A0A088ARE4 A0A2A3EE79 A0A232EM52 A0A0M9A6E1 K7IM19 A0A0L7RIJ3 A0A1B6J1A6 Q16SB9 A0A023EVG3 E9J4H9 A0A182GE15 A0A158NM78 A0A151JWD3 B0WLL0 F4WK04 A0A1Q3F0M9 E0VL42 E2A8Z0 A0A026WHT9 A0A151IRI2 A0A151X2E0 A0A151IE81 A0A0J7L327 A0A195BYV4 A0A182RK53 E2B7T3 A0A0C9QK38 U5EXW5 A0A336K919 A0A182LUB3 A0A182WFG0 A0A1B6F7D1 A0A182YSB3 A0A182JX61 A0A182X5W3 W5JJ08 A0A182SCW5 A0A182VC82 A0A182KN71 A0A182IBJ6 A0A2M4BGD2 A0A2M4BG96 A0A2M4BGA4 A0A182TP29 Q7Q8G9 A0A182P488 A0A2M4AIY1 A0A182ISB0 A0A2M4AJ56 A0A210PH49 A0A182NGP2 A0A3Q0J3U9 A0A084W7N9 A0A182Q1M4 A0A2C9K596 A0A1S3J3I5 A0A2R2MPD2 A0A1S3J4Q2 A0A0B7BGF8 A0A1J1HST2 A0A1B0CYM6 K1Q5G9 A0A0L0C1Y7 A0A1B6DVJ7 V4ATI1 A0A1L8DST4 A0A0T6B393 A0A2T7NHS4 A0A1A9ZX08 A0A2B4RUV6 A0A1W4WFV1 A0A1Y1KF70 A0A1B0G3Y6 A0A1A9XIL0 A0A1B0BCP7 A0A3M6U7E3 A0A0N8DRQ8 A0A1I8PHE9 A0A0P5E456 A0A0P4XPT0 A0A0N8CL88 A0A0P5ZWY1 A0A0P4YU31 A0A0P6E5M0 A0A165A5B7 A0A1I8MXD0 A0A0P5XLD6 A0A1A9XPX7 A0A1A9VN27 A0A1B0G2L0
Pubmed
19121390
26354079
22118469
23622113
24845553
28648823
+ More
20075255 17510324 24945155 21282665 26483478 21347285 21719571 20566863 20798317 24508170 30249741 25244985 20920257 23761445 20966253 12364791 14747013 17210077 28812685 24438588 15562597 22992520 26108605 23254933 28004739 30382153 25315136
20075255 17510324 24945155 21282665 26483478 21347285 21719571 20566863 20798317 24508170 30249741 25244985 20920257 23761445 20966253 12364791 14747013 17210077 28812685 24438588 15562597 22992520 26108605 23254933 28004739 30382153 25315136
EMBL
BABH01016361
ODYU01001443
SOQ37643.1
KQ459252
KPJ02113.1
AGBW02014303
+ More
OWR41906.1 RSAL01000107 RVE47291.1 GAIX01004929 JAA87631.1 NWSH01003386 PCG66409.1 NEVH01014359 PNF27775.1 KK852879 KDR14472.1 KZ288282 PBC29592.1 NNAY01003438 OXU19417.1 KQ435726 KOX78175.1 KQ414584 KOC70649.1 GECU01014816 JAS92890.1 CH477677 EAT37350.1 GAPW01000470 JAC13128.1 GL768105 EFZ12246.1 JXUM01056951 KQ561933 KXJ77119.1 ADTU01020341 KQ981652 KYN38631.1 DS231987 EDS30540.1 GL888190 EGI65454.1 GFDL01013939 JAV21106.1 DS235269 EEB14098.1 GL437703 EFN70089.1 KK107238 QOIP01000007 EZA54639.1 RLU20620.1 KQ981126 KYN09312.1 KQ982584 KYQ54419.1 KQ977890 KYM98974.1 LBMM01001137 KMQ96839.1 KQ976395 KYM93103.1 GL446209 EFN88226.1 GBYB01015168 JAG84935.1 GANO01000712 JAB59159.1 UFQS01000126 UFQT01000126 SSX00138.1 SSX20518.1 AXCM01002194 GECZ01027853 GECZ01023694 JAS41916.1 JAS46075.1 ADMH02001096 ETN64121.1 APCN01000707 GGFJ01002936 MBW52077.1 GGFJ01002934 MBW52075.1 GGFJ01002933 MBW52074.1 AAAB01008944 EAA10202.3 GGFK01007410 MBW40731.1 GGFK01007451 MBW40772.1 NEDP02076706 OWF35811.1 ATLV01021268 KE525315 KFB46233.1 AXCN02002080 HACG01044536 CEK91401.1 CVRI01000020 CRK91061.1 AJVK01009248 JH816675 EKC24155.1 JRES01000997 KNC26267.1 GEDC01007604 JAS29694.1 KB201262 ESO98215.1 GFDF01004660 JAV09424.1 LJIG01016014 KRT81872.1 PZQS01000012 PVD20729.1 LSMT01000246 PFX22234.1 GEZM01085156 JAV60173.1 CCAG010017310 JXJN01012081 RCHS01002146 RMX49378.1 GDIP01007383 JAM96332.1 GDIP01146787 JAJ76615.1 GDIP01239485 GDIP01230635 GDIP01071953 JAI83916.1 GDIP01120743 GDIP01082681 JAL82971.1 GDIP01124894 GDIP01038205 JAM65510.1 GDIP01222325 JAJ01077.1 GDIQ01068658 JAN26079.1 LRGB01000642 KZS17193.1 GDIP01071952 JAM31763.1 CCAG010007952
OWR41906.1 RSAL01000107 RVE47291.1 GAIX01004929 JAA87631.1 NWSH01003386 PCG66409.1 NEVH01014359 PNF27775.1 KK852879 KDR14472.1 KZ288282 PBC29592.1 NNAY01003438 OXU19417.1 KQ435726 KOX78175.1 KQ414584 KOC70649.1 GECU01014816 JAS92890.1 CH477677 EAT37350.1 GAPW01000470 JAC13128.1 GL768105 EFZ12246.1 JXUM01056951 KQ561933 KXJ77119.1 ADTU01020341 KQ981652 KYN38631.1 DS231987 EDS30540.1 GL888190 EGI65454.1 GFDL01013939 JAV21106.1 DS235269 EEB14098.1 GL437703 EFN70089.1 KK107238 QOIP01000007 EZA54639.1 RLU20620.1 KQ981126 KYN09312.1 KQ982584 KYQ54419.1 KQ977890 KYM98974.1 LBMM01001137 KMQ96839.1 KQ976395 KYM93103.1 GL446209 EFN88226.1 GBYB01015168 JAG84935.1 GANO01000712 JAB59159.1 UFQS01000126 UFQT01000126 SSX00138.1 SSX20518.1 AXCM01002194 GECZ01027853 GECZ01023694 JAS41916.1 JAS46075.1 ADMH02001096 ETN64121.1 APCN01000707 GGFJ01002936 MBW52077.1 GGFJ01002934 MBW52075.1 GGFJ01002933 MBW52074.1 AAAB01008944 EAA10202.3 GGFK01007410 MBW40731.1 GGFK01007451 MBW40772.1 NEDP02076706 OWF35811.1 ATLV01021268 KE525315 KFB46233.1 AXCN02002080 HACG01044536 CEK91401.1 CVRI01000020 CRK91061.1 AJVK01009248 JH816675 EKC24155.1 JRES01000997 KNC26267.1 GEDC01007604 JAS29694.1 KB201262 ESO98215.1 GFDF01004660 JAV09424.1 LJIG01016014 KRT81872.1 PZQS01000012 PVD20729.1 LSMT01000246 PFX22234.1 GEZM01085156 JAV60173.1 CCAG010017310 JXJN01012081 RCHS01002146 RMX49378.1 GDIP01007383 JAM96332.1 GDIP01146787 JAJ76615.1 GDIP01239485 GDIP01230635 GDIP01071953 JAI83916.1 GDIP01120743 GDIP01082681 JAL82971.1 GDIP01124894 GDIP01038205 JAM65510.1 GDIP01222325 JAJ01077.1 GDIQ01068658 JAN26079.1 LRGB01000642 KZS17193.1 GDIP01071952 JAM31763.1 CCAG010007952
Proteomes
UP000005204
UP000053268
UP000007151
UP000283053
UP000218220
UP000235965
+ More
UP000027135 UP000005203 UP000242457 UP000215335 UP000053105 UP000002358 UP000053825 UP000008820 UP000069940 UP000249989 UP000005205 UP000078541 UP000002320 UP000007755 UP000009046 UP000000311 UP000053097 UP000279307 UP000078492 UP000075809 UP000078542 UP000036403 UP000078540 UP000075900 UP000008237 UP000075883 UP000075920 UP000076408 UP000075881 UP000076407 UP000000673 UP000075901 UP000075903 UP000075882 UP000075840 UP000075902 UP000007062 UP000075885 UP000075880 UP000242188 UP000075884 UP000079169 UP000030765 UP000075886 UP000076420 UP000085678 UP000183832 UP000092462 UP000005408 UP000037069 UP000030746 UP000245119 UP000092445 UP000225706 UP000192223 UP000092444 UP000092443 UP000092460 UP000275408 UP000095300 UP000076858 UP000095301 UP000078200
UP000027135 UP000005203 UP000242457 UP000215335 UP000053105 UP000002358 UP000053825 UP000008820 UP000069940 UP000249989 UP000005205 UP000078541 UP000002320 UP000007755 UP000009046 UP000000311 UP000053097 UP000279307 UP000078492 UP000075809 UP000078542 UP000036403 UP000078540 UP000075900 UP000008237 UP000075883 UP000075920 UP000076408 UP000075881 UP000076407 UP000000673 UP000075901 UP000075903 UP000075882 UP000075840 UP000075902 UP000007062 UP000075885 UP000075880 UP000242188 UP000075884 UP000079169 UP000030765 UP000075886 UP000076420 UP000085678 UP000183832 UP000092462 UP000005408 UP000037069 UP000030746 UP000245119 UP000092445 UP000225706 UP000192223 UP000092444 UP000092443 UP000092460 UP000275408 UP000095300 UP000076858 UP000095301 UP000078200
PRIDE
Pfam
Interpro
IPR023318
Ub_act_enz_dom_a_sf
+ More
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR028077 UAE_UbL_dom
IPR019572 UBA_E1_Cys
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR030661 Uba2
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR032426 UBA2_C
IPR011705 BACK
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR028077 UAE_UbL_dom
IPR019572 UBA_E1_Cys
IPR035985 Ubiquitin-activating_enz
IPR000594 ThiF_NAD_FAD-bd
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR030661 Uba2
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR032426 UBA2_C
IPR011705 BACK
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
Gene 3D
CDD
ProteinModelPortal
H9JRF6
A0A2H1V9X0
A0A194Q9D4
A0A212EK97
A0A3S2LYU5
S4P6Q4
+ More
A0A2A4J3G1 A0A2J7QGR2 A0A067R5B3 A0A088ARE4 A0A2A3EE79 A0A232EM52 A0A0M9A6E1 K7IM19 A0A0L7RIJ3 A0A1B6J1A6 Q16SB9 A0A023EVG3 E9J4H9 A0A182GE15 A0A158NM78 A0A151JWD3 B0WLL0 F4WK04 A0A1Q3F0M9 E0VL42 E2A8Z0 A0A026WHT9 A0A151IRI2 A0A151X2E0 A0A151IE81 A0A0J7L327 A0A195BYV4 A0A182RK53 E2B7T3 A0A0C9QK38 U5EXW5 A0A336K919 A0A182LUB3 A0A182WFG0 A0A1B6F7D1 A0A182YSB3 A0A182JX61 A0A182X5W3 W5JJ08 A0A182SCW5 A0A182VC82 A0A182KN71 A0A182IBJ6 A0A2M4BGD2 A0A2M4BG96 A0A2M4BGA4 A0A182TP29 Q7Q8G9 A0A182P488 A0A2M4AIY1 A0A182ISB0 A0A2M4AJ56 A0A210PH49 A0A182NGP2 A0A3Q0J3U9 A0A084W7N9 A0A182Q1M4 A0A2C9K596 A0A1S3J3I5 A0A2R2MPD2 A0A1S3J4Q2 A0A0B7BGF8 A0A1J1HST2 A0A1B0CYM6 K1Q5G9 A0A0L0C1Y7 A0A1B6DVJ7 V4ATI1 A0A1L8DST4 A0A0T6B393 A0A2T7NHS4 A0A1A9ZX08 A0A2B4RUV6 A0A1W4WFV1 A0A1Y1KF70 A0A1B0G3Y6 A0A1A9XIL0 A0A1B0BCP7 A0A3M6U7E3 A0A0N8DRQ8 A0A1I8PHE9 A0A0P5E456 A0A0P4XPT0 A0A0N8CL88 A0A0P5ZWY1 A0A0P4YU31 A0A0P6E5M0 A0A165A5B7 A0A1I8MXD0 A0A0P5XLD6 A0A1A9XPX7 A0A1A9VN27 A0A1B0G2L0
A0A2A4J3G1 A0A2J7QGR2 A0A067R5B3 A0A088ARE4 A0A2A3EE79 A0A232EM52 A0A0M9A6E1 K7IM19 A0A0L7RIJ3 A0A1B6J1A6 Q16SB9 A0A023EVG3 E9J4H9 A0A182GE15 A0A158NM78 A0A151JWD3 B0WLL0 F4WK04 A0A1Q3F0M9 E0VL42 E2A8Z0 A0A026WHT9 A0A151IRI2 A0A151X2E0 A0A151IE81 A0A0J7L327 A0A195BYV4 A0A182RK53 E2B7T3 A0A0C9QK38 U5EXW5 A0A336K919 A0A182LUB3 A0A182WFG0 A0A1B6F7D1 A0A182YSB3 A0A182JX61 A0A182X5W3 W5JJ08 A0A182SCW5 A0A182VC82 A0A182KN71 A0A182IBJ6 A0A2M4BGD2 A0A2M4BG96 A0A2M4BGA4 A0A182TP29 Q7Q8G9 A0A182P488 A0A2M4AIY1 A0A182ISB0 A0A2M4AJ56 A0A210PH49 A0A182NGP2 A0A3Q0J3U9 A0A084W7N9 A0A182Q1M4 A0A2C9K596 A0A1S3J3I5 A0A2R2MPD2 A0A1S3J4Q2 A0A0B7BGF8 A0A1J1HST2 A0A1B0CYM6 K1Q5G9 A0A0L0C1Y7 A0A1B6DVJ7 V4ATI1 A0A1L8DST4 A0A0T6B393 A0A2T7NHS4 A0A1A9ZX08 A0A2B4RUV6 A0A1W4WFV1 A0A1Y1KF70 A0A1B0G3Y6 A0A1A9XIL0 A0A1B0BCP7 A0A3M6U7E3 A0A0N8DRQ8 A0A1I8PHE9 A0A0P5E456 A0A0P4XPT0 A0A0N8CL88 A0A0P5ZWY1 A0A0P4YU31 A0A0P6E5M0 A0A165A5B7 A0A1I8MXD0 A0A0P5XLD6 A0A1A9XPX7 A0A1A9VN27 A0A1B0G2L0
PDB
3KYD
E-value=2.91026e-163,
Score=1478
Ontologies
GO
PANTHER
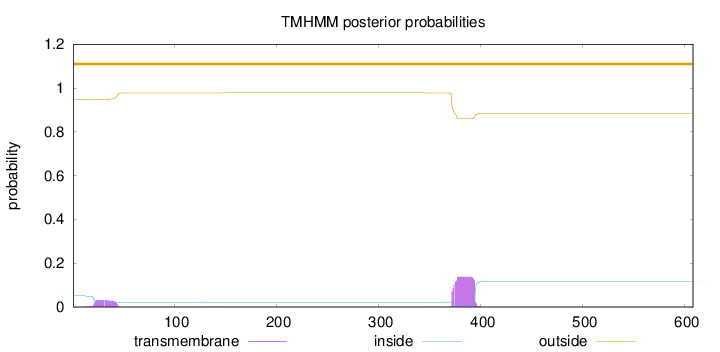
Topology
Length:
608
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.71592
Exp number, first 60 AAs:
0.70552
Total prob of N-in:
0.05175
outside
1 - 608
Population Genetic Test Statistics
Pi
5.226232
Theta
14.376044
Tajima's D
-1.618822
CLR
1.291564
CSRT
0.0439978001099945
Interpretation
Uncertain
