Pre Gene Modal
BGIBMGA012038
Annotation
putative_o-sialoglycoprotein_endopeptidase_[Danaus_plexippus]
Full name
Probable tRNA N6-adenosine threonylcarbamoyltransferase
Alternative Name
t(6)A37 threonylcarbamoyladenosine biosynthesis protein
tRNA threonylcarbamoyladenosine biosynthesis protein
t(6)A37 threonylcarbamoyladenosine biosynthesis protein CSON001408
tRNA threonylcarbamoyladenosine biosynthesis protein CSON001408
t(6)A37 threonylcarbamoyladenosine biosynthesis protein CSON001875
tRNA threonylcarbamoyladenosine biosynthesis protein CSON001875
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 5573005
tRNA threonylcarbamoyladenosine biosynthesis protein 5573005
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 6042337
tRNA threonylcarbamoyladenosine biosynthesis protein 6042337
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 1270132
tRNA threonylcarbamoyladenosine biosynthesis protein 1270132
t(6)A37 threonylcarbamoyladenosine biosynthesis protein osgep_2
tRNA threonylcarbamoyladenosine biosynthesis protein osgep_2
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 8238097
tRNA threonylcarbamoyladenosine biosynthesis protein 8238097
t(6)A37 threonylcarbamoyladenosine biosynthesis protein OSGEP
tRNA threonylcarbamoyladenosine biosynthesis protein OSGEP
O-sialoglycoprotein endopeptidase
t(6)A37 threonylcarbamoyladenosine biosynthesis protein ACYPI004911
tRNA threonylcarbamoyladenosine biosynthesis protein ACYPI004911
t(6)A37 threonylcarbamoyladenosine biosynthesis protein osgep
tRNA threonylcarbamoyladenosine biosynthesis protein osgep
t(6)A37 threonylcarbamoyladenosine biosynthesis protein LOC106176207
tRNA threonylcarbamoyladenosine biosynthesis protein LOC106176207
t(6)A37 threonylcarbamoyladenosine biosynthesis protein CG4933_0
tRNA threonylcarbamoyladenosine biosynthesis protein CG4933_0
N6-L-threonylcarbamoyladenine synthase
t(6)A37 threonylcarbamoyladenosine biosynthesis protein LOC108737171
tRNA threonylcarbamoyladenosine biosynthesis protein LOC108737171
t(6)A37 threonylcarbamoyladenosine biosynthesis protein LOC106576589
tRNA threonylcarbamoyladenosine biosynthesis protein LOC106576589
tRNA threonylcarbamoyladenosine biosynthesis protein
t(6)A37 threonylcarbamoyladenosine biosynthesis protein CSON001408
tRNA threonylcarbamoyladenosine biosynthesis protein CSON001408
t(6)A37 threonylcarbamoyladenosine biosynthesis protein CSON001875
tRNA threonylcarbamoyladenosine biosynthesis protein CSON001875
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 5573005
tRNA threonylcarbamoyladenosine biosynthesis protein 5573005
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 6042337
tRNA threonylcarbamoyladenosine biosynthesis protein 6042337
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 1270132
tRNA threonylcarbamoyladenosine biosynthesis protein 1270132
t(6)A37 threonylcarbamoyladenosine biosynthesis protein osgep_2
tRNA threonylcarbamoyladenosine biosynthesis protein osgep_2
t(6)A37 threonylcarbamoyladenosine biosynthesis protein 8238097
tRNA threonylcarbamoyladenosine biosynthesis protein 8238097
t(6)A37 threonylcarbamoyladenosine biosynthesis protein OSGEP
tRNA threonylcarbamoyladenosine biosynthesis protein OSGEP
O-sialoglycoprotein endopeptidase
t(6)A37 threonylcarbamoyladenosine biosynthesis protein ACYPI004911
tRNA threonylcarbamoyladenosine biosynthesis protein ACYPI004911
t(6)A37 threonylcarbamoyladenosine biosynthesis protein osgep
tRNA threonylcarbamoyladenosine biosynthesis protein osgep
t(6)A37 threonylcarbamoyladenosine biosynthesis protein LOC106176207
tRNA threonylcarbamoyladenosine biosynthesis protein LOC106176207
t(6)A37 threonylcarbamoyladenosine biosynthesis protein CG4933_0
tRNA threonylcarbamoyladenosine biosynthesis protein CG4933_0
N6-L-threonylcarbamoyladenine synthase
t(6)A37 threonylcarbamoyladenosine biosynthesis protein LOC108737171
tRNA threonylcarbamoyladenosine biosynthesis protein LOC108737171
t(6)A37 threonylcarbamoyladenosine biosynthesis protein LOC106576589
tRNA threonylcarbamoyladenosine biosynthesis protein LOC106576589
Location in the cell
Cytoplasmic Reliability : 3.014
Sequence
CDS
ATGGTAGTAGCAATAGGATTTGAGGGAAGTGCAAACAAACTGGGAATTGGTATAGTTAAAGATGGAGAAATCTTGGCCAATTGCAGAAGAACGTACATAACCCCACCCGGTGAAGGGTTTCTTCCTAGAGAAACTGCGGAGCATCATCAACAAAACATTCATGAAGTATTGCAAGAAGCTTTGGACCAGTCTGGTCTCAATCCAGATGAAATTGACGTTGTCTGTTATACCAAAGGACCTGGTATGGGGGCGCCATTGATGGTGTGTGCAATTGTTGCCAGAACATGTGCTAAACTATGGAAAAAACCTATTTACGGTGTAAATCATTGTATTGGACATATAGAAATGGGAAGGTTGATAACCAAAGCAAATAATCCAACAGTGCTATATGTGAGTGGAGGGAATACACAAATTATAGCATATTCAAGAAAAAGATACAGGATATTTGGTGAAACTATTGATATAGCTGTTGGCAACTGCTTAGATAGGTTTGCTAGAGTTTTGAAATTGTCTAATGCTCCAAGTCCAGGTTATAACATAGAACAAGCTGCTAAAAAAGGAAAAAAGTATTTGCCCTTACCATATTGTGTTAAAGGAATGGACGTAAGTTTTTCTGGTATCCTATCGTACATGGAGGATAAAATTGACGAATTATTGAAAGAATATACTCCGGAAGACTTATGTTATTCTTTGCAAGAAACTGTTTTTGCAATGCTAGTGGAGATCACTGAACGAGCTATGGCTCACTGCGGTTCTGATGAGGTATTAATAGTAGGTGGAGTTGGGTGCAATGAGCGACTTCAAGAGATGATGGGAGTAATGTGTCATGAACGAGAGGCTAAGATATTTGCTACCGATGAGAGATTTTGCATAGATAATGGAGTGATGATAGCATATGCTGGTGCCCTTGCTCATGCTTCTGGCACTCGCATGGATTTCAAAGATGCAACAATTACACAAAGATATAGGACAGATGATGTTCTTGTTACGTGGCGAGATGATTAA
Protein
MVVAIGFEGSANKLGIGIVKDGEILANCRRTYITPPGEGFLPRETAEHHQQNIHEVLQEALDQSGLNPDEIDVVCYTKGPGMGAPLMVCAIVARTCAKLWKKPIYGVNHCIGHIEMGRLITKANNPTVLYVSGGNTQIIAYSRKRYRIFGETIDIAVGNCLDRFARVLKLSNAPSPGYNIEQAAKKGKKYLPLPYCVKGMDVSFSGILSYMEDKIDELLKEYTPEDLCYSLQETVFAMLVEITERAMAHCGSDEVLIVGGVGCNERLQEMMGVMCHEREAKIFATDERFCIDNGVMIAYAGALAHASGTRMDFKDATITQRYRTDDVLVTWRDD
Summary
Description
Component of the EKC/KEOPS complex that is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. The complex is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37. Likely plays a direct catalytic role in this reaction, but requires other protein(s) of the complex to fulfill this activity.
Component of the EKC/KEOPS complex that is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. The complex is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37. OSGEP likely plays a direct catalytic role in this reaction, but requires other protein(s) of the complex to fulfill this activity.
Component of the EKC/KEOPS complex that is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. The complex is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37. Osgep likely plays a direct catalytic role in this reaction, but requires other protein(s) of the complex to fulfill this activity.
Component of the EKC/KEOPS complex that is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. The complex is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37. OSGEP likely plays a direct catalytic role in this reaction, but requires other protein(s) of the complex to fulfill this activity.
Component of the EKC/KEOPS complex that is required for the formation of a threonylcarbamoyl group on adenosine at position 37 (t(6)A37) in tRNAs that read codons beginning with adenine. The complex is probably involved in the transfer of the threonylcarbamoyl moiety of threonylcarbamoyl-AMP (TC-AMP) to the N6 group of A37. Osgep likely plays a direct catalytic role in this reaction, but requires other protein(s) of the complex to fulfill this activity.
Catalytic Activity
adenosine(37) in tRNA + L-threonylcarbamoyladenylate = AMP + H(+) + N(6)-L-threonylcarbamoyladenosine(37) in tRNA
Cofactor
a divalent metal cation
Subunit
Component of the EKC/KEOPS complex; the whole complex dimerizes.
Component of the EKC/KEOPS complex composed of at least TP53RK, TPRKB, OSGEP and LAGE3; the whole complex dimerizes.
Component of the EKC/KEOPS complex composed of at least tp53rk, tprkb, osgep and lage3; the whole complex dimerizes.
Component of the EKC/KEOPS complex composed of at least TP53RK, TPRKB, OSGEP and LAGE3; the whole complex dimerizes.
Component of the EKC/KEOPS complex composed of at least tp53rk, tprkb, osgep and lage3; the whole complex dimerizes.
Similarity
Belongs to the KAE1 / TsaD family.
Keywords
Acyltransferase
Complete proteome
Cytoplasm
Metal-binding
Nucleus
Reference proteome
Transferase
tRNA processing
Feature
chain Probable tRNA N6-adenosine threonylcarbamoyltransferase
Uniprot
A0A3S2TIX0
A0A212F800
A0A2A4J5L9
A0A2H1VA26
S4PF58
A0A194QQ51
+ More
A0A336L7E3 A0A336L8H4 A0A1L8DTJ2 D6WGD4 A0A1B6LLM1 N6URV1 A0A1B6IXS4 U4UYN0 A0A1B6FX76 Q17JQ2 A0A1Q3F2H3 Q17JQ4 A0A182G8M0 A0A1S4F082 J3JXV9 B0WRY1 A0A1B6E6R6 A0A182N996 A0A1Y1L519 A0A182TT82 Q7QIM5 A0A2J7QBB4 A0A182VIC0 A0A182L3L9 A0A182I6S2 A0A182WWJ1 E0VCY3 A0A131XE09 W8C880 A0A1I8JUP0 A0A182QUX9 A0A3B3DUR2 A0A2M4BST6 A0A0K8R5M8 A0A182IR96 A0A182Y2C8 C4WVI6 A0A2P8XPS1 A0A2M4BST1 W5JPN9 A0A2M4ANZ2 A0A2M3Z9Y1 A0A3B4YWS2 A0A3Q4MCJ4 A0A3Q3JJ62 A0A2U9B1H6 A0A3P8TVH2 A0A182SXW4 A0A182PII1 I3K266 A0A1S3JUE1 A0A3Q3JCH8 A0A182JP77 Q5RHZ6 A0A3P8QCK5 A0A3B4H8Y6 A0A3Q3BRA2 A0A3P9C9B6 Q4SY05 A0A3B4UVE1 A0A131Z5L4 A0A067R3I0 A0A3B4YJJ8 A0A034VQT7 A0A224YIZ2 L7MF93 A0A1L8I071 A0A3Q2XSL8 A0A0S7MDV1 A0A3P9AJ18 A0A2D0T2K0 E3TBP5 A0A1E1XQA4 A0A0K8V745 A0A146N6M5 A0A232FHF2 A0A154PDK5 A0A087XE15 A0A3P9PGV7 A0A3Q1IID3 Q7SYR1 A0A1W4WZ88 A0A3B3U9G9 A0A146XJ08 A0A3B3R4S0 A0A1S3N1L8 A0A147AIE0 A0A3Q3T2J8 A0A1J1J4I1 A0A3Q2EIG5 A0A3B5R332 Q0V9I9 G3MH93 A0A0N7ZDA9
A0A336L7E3 A0A336L8H4 A0A1L8DTJ2 D6WGD4 A0A1B6LLM1 N6URV1 A0A1B6IXS4 U4UYN0 A0A1B6FX76 Q17JQ2 A0A1Q3F2H3 Q17JQ4 A0A182G8M0 A0A1S4F082 J3JXV9 B0WRY1 A0A1B6E6R6 A0A182N996 A0A1Y1L519 A0A182TT82 Q7QIM5 A0A2J7QBB4 A0A182VIC0 A0A182L3L9 A0A182I6S2 A0A182WWJ1 E0VCY3 A0A131XE09 W8C880 A0A1I8JUP0 A0A182QUX9 A0A3B3DUR2 A0A2M4BST6 A0A0K8R5M8 A0A182IR96 A0A182Y2C8 C4WVI6 A0A2P8XPS1 A0A2M4BST1 W5JPN9 A0A2M4ANZ2 A0A2M3Z9Y1 A0A3B4YWS2 A0A3Q4MCJ4 A0A3Q3JJ62 A0A2U9B1H6 A0A3P8TVH2 A0A182SXW4 A0A182PII1 I3K266 A0A1S3JUE1 A0A3Q3JCH8 A0A182JP77 Q5RHZ6 A0A3P8QCK5 A0A3B4H8Y6 A0A3Q3BRA2 A0A3P9C9B6 Q4SY05 A0A3B4UVE1 A0A131Z5L4 A0A067R3I0 A0A3B4YJJ8 A0A034VQT7 A0A224YIZ2 L7MF93 A0A1L8I071 A0A3Q2XSL8 A0A0S7MDV1 A0A3P9AJ18 A0A2D0T2K0 E3TBP5 A0A1E1XQA4 A0A0K8V745 A0A146N6M5 A0A232FHF2 A0A154PDK5 A0A087XE15 A0A3P9PGV7 A0A3Q1IID3 Q7SYR1 A0A1W4WZ88 A0A3B3U9G9 A0A146XJ08 A0A3B3R4S0 A0A1S3N1L8 A0A147AIE0 A0A3Q3T2J8 A0A1J1J4I1 A0A3Q2EIG5 A0A3B5R332 Q0V9I9 G3MH93 A0A0N7ZDA9
EC Number
2.3.1.234
Pubmed
22118469
23622113
26354079
18362917
19820115
23537049
+ More
17510324 26483478 22516182 28004739 12364791 14747013 17210077 20966253 20566863 28049606 24495485 29451363 25244985 29403074 20920257 23761445 25186727 23594743 29346415 28805828 15496914 26830274 24845553 25348373 28797301 25576852 27762356 25069045 20634964 29209593 28648823 29240929 23542700 20431018 22216098
17510324 26483478 22516182 28004739 12364791 14747013 17210077 20966253 20566863 28049606 24495485 29451363 25244985 29403074 20920257 23761445 25186727 23594743 29346415 28805828 15496914 26830274 24845553 25348373 28797301 25576852 27762356 25069045 20634964 29209593 28648823 29240929 23542700 20431018 22216098
EMBL
RSAL01000107
RVE47286.1
AGBW02009812
OWR49860.1
NWSH01003235
PCG66713.1
+ More
ODYU01001443 SOQ37646.1 GAIX01004202 JAA88358.1 KQ461185 KPJ07489.1 UFQS01001209 UFQT01001209 SSX09785.1 SSX29508.1 UFQS01001301 UFQT01001301 SSX10185.1 SSX29906.1 GFDF01004357 JAV09727.1 KQ971329 EFA01122.1 GEBQ01015336 JAT24641.1 APGK01019136 KB740092 ENN81462.1 GECU01015955 JAS91751.1 KB632429 ERL95430.1 GECZ01014974 JAS54795.1 CH477231 EAT46922.1 GFDL01013316 JAV21729.1 EAT46920.1 JXUM01008040 KQ560252 KXJ83547.1 BT128081 AEE63042.1 DS232062 EDS33591.1 GEDC01028342 GEDC01017293 GEDC01003675 JAS08956.1 JAS20005.1 JAS33623.1 GEZM01066418 JAV67898.1 AAAB01008807 EAA04730.2 NEVH01016301 PNF25867.1 APCN01003693 DS235068 EEB11239.1 GEFH01004705 JAP63876.1 GAMC01000082 JAC06474.1 AXCN02000558 GGFJ01007006 MBW56147.1 GADI01007336 JAA66472.1 ABLF02038097 AK341535 BAH71906.1 PYGN01001569 PSN33990.1 GGFJ01007005 MBW56146.1 ADMH02000528 ETN66096.1 GGFK01009174 MBW42495.1 GGFM01004583 MBW25334.1 CP026244 AWO97792.1 AERX01022926 AERX01022927 BX323558 BC093366 BC164136 CAAE01012247 CAF94477.1 GEDV01002497 JAP86060.1 KK852729 KDR17566.1 GAKP01014128 JAC44824.1 GFPF01004553 MAA15699.1 GACK01003175 JAA61859.1 CM004466 OCU01558.1 GBYX01067356 JAO99900.1 GU587770 ADO27731.1 GFAA01001970 JAU01465.1 GDHF01017592 GDHF01009827 JAI34722.1 JAI42487.1 GCES01158818 JAQ27504.1 NNAY01000242 OXU29767.1 KQ434870 KZC09464.1 AYCK01014076 BC054300 GCES01044293 JAR42030.1 GCES01007939 JAR78384.1 CVRI01000070 CRL07317.1 AAMC01032139 AAMC01032140 AAMC01032141 BC121530 AAI21531.1 JO841244 AEO32861.1 GDRN01047212 JAI66736.1
ODYU01001443 SOQ37646.1 GAIX01004202 JAA88358.1 KQ461185 KPJ07489.1 UFQS01001209 UFQT01001209 SSX09785.1 SSX29508.1 UFQS01001301 UFQT01001301 SSX10185.1 SSX29906.1 GFDF01004357 JAV09727.1 KQ971329 EFA01122.1 GEBQ01015336 JAT24641.1 APGK01019136 KB740092 ENN81462.1 GECU01015955 JAS91751.1 KB632429 ERL95430.1 GECZ01014974 JAS54795.1 CH477231 EAT46922.1 GFDL01013316 JAV21729.1 EAT46920.1 JXUM01008040 KQ560252 KXJ83547.1 BT128081 AEE63042.1 DS232062 EDS33591.1 GEDC01028342 GEDC01017293 GEDC01003675 JAS08956.1 JAS20005.1 JAS33623.1 GEZM01066418 JAV67898.1 AAAB01008807 EAA04730.2 NEVH01016301 PNF25867.1 APCN01003693 DS235068 EEB11239.1 GEFH01004705 JAP63876.1 GAMC01000082 JAC06474.1 AXCN02000558 GGFJ01007006 MBW56147.1 GADI01007336 JAA66472.1 ABLF02038097 AK341535 BAH71906.1 PYGN01001569 PSN33990.1 GGFJ01007005 MBW56146.1 ADMH02000528 ETN66096.1 GGFK01009174 MBW42495.1 GGFM01004583 MBW25334.1 CP026244 AWO97792.1 AERX01022926 AERX01022927 BX323558 BC093366 BC164136 CAAE01012247 CAF94477.1 GEDV01002497 JAP86060.1 KK852729 KDR17566.1 GAKP01014128 JAC44824.1 GFPF01004553 MAA15699.1 GACK01003175 JAA61859.1 CM004466 OCU01558.1 GBYX01067356 JAO99900.1 GU587770 ADO27731.1 GFAA01001970 JAU01465.1 GDHF01017592 GDHF01009827 JAI34722.1 JAI42487.1 GCES01158818 JAQ27504.1 NNAY01000242 OXU29767.1 KQ434870 KZC09464.1 AYCK01014076 BC054300 GCES01044293 JAR42030.1 GCES01007939 JAR78384.1 CVRI01000070 CRL07317.1 AAMC01032139 AAMC01032140 AAMC01032141 BC121530 AAI21531.1 JO841244 AEO32861.1 GDRN01047212 JAI66736.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053240
UP000007266
UP000019118
+ More
UP000030742 UP000008820 UP000069940 UP000249989 UP000002320 UP000075884 UP000075902 UP000007062 UP000235965 UP000075903 UP000075882 UP000075840 UP000076407 UP000009046 UP000075900 UP000075886 UP000261560 UP000075880 UP000076408 UP000007819 UP000245037 UP000000673 UP000261400 UP000261580 UP000261600 UP000246464 UP000265080 UP000075901 UP000075885 UP000005207 UP000085678 UP000075881 UP000000437 UP000265100 UP000261460 UP000264840 UP000265160 UP000007303 UP000261420 UP000027135 UP000261360 UP000186698 UP000264820 UP000265140 UP000221080 UP000215335 UP000076502 UP000028760 UP000242638 UP000265040 UP000192223 UP000261500 UP000265000 UP000261540 UP000087266 UP000261640 UP000183832 UP000265020 UP000002852 UP000008143
UP000030742 UP000008820 UP000069940 UP000249989 UP000002320 UP000075884 UP000075902 UP000007062 UP000235965 UP000075903 UP000075882 UP000075840 UP000076407 UP000009046 UP000075900 UP000075886 UP000261560 UP000075880 UP000076408 UP000007819 UP000245037 UP000000673 UP000261400 UP000261580 UP000261600 UP000246464 UP000265080 UP000075901 UP000075885 UP000005207 UP000085678 UP000075881 UP000000437 UP000265100 UP000261460 UP000264840 UP000265160 UP000007303 UP000261420 UP000027135 UP000261360 UP000186698 UP000264820 UP000265140 UP000221080 UP000215335 UP000076502 UP000028760 UP000242638 UP000265040 UP000192223 UP000261500 UP000265000 UP000261540 UP000087266 UP000261640 UP000183832 UP000265020 UP000002852 UP000008143
Interpro
ProteinModelPortal
A0A3S2TIX0
A0A212F800
A0A2A4J5L9
A0A2H1VA26
S4PF58
A0A194QQ51
+ More
A0A336L7E3 A0A336L8H4 A0A1L8DTJ2 D6WGD4 A0A1B6LLM1 N6URV1 A0A1B6IXS4 U4UYN0 A0A1B6FX76 Q17JQ2 A0A1Q3F2H3 Q17JQ4 A0A182G8M0 A0A1S4F082 J3JXV9 B0WRY1 A0A1B6E6R6 A0A182N996 A0A1Y1L519 A0A182TT82 Q7QIM5 A0A2J7QBB4 A0A182VIC0 A0A182L3L9 A0A182I6S2 A0A182WWJ1 E0VCY3 A0A131XE09 W8C880 A0A1I8JUP0 A0A182QUX9 A0A3B3DUR2 A0A2M4BST6 A0A0K8R5M8 A0A182IR96 A0A182Y2C8 C4WVI6 A0A2P8XPS1 A0A2M4BST1 W5JPN9 A0A2M4ANZ2 A0A2M3Z9Y1 A0A3B4YWS2 A0A3Q4MCJ4 A0A3Q3JJ62 A0A2U9B1H6 A0A3P8TVH2 A0A182SXW4 A0A182PII1 I3K266 A0A1S3JUE1 A0A3Q3JCH8 A0A182JP77 Q5RHZ6 A0A3P8QCK5 A0A3B4H8Y6 A0A3Q3BRA2 A0A3P9C9B6 Q4SY05 A0A3B4UVE1 A0A131Z5L4 A0A067R3I0 A0A3B4YJJ8 A0A034VQT7 A0A224YIZ2 L7MF93 A0A1L8I071 A0A3Q2XSL8 A0A0S7MDV1 A0A3P9AJ18 A0A2D0T2K0 E3TBP5 A0A1E1XQA4 A0A0K8V745 A0A146N6M5 A0A232FHF2 A0A154PDK5 A0A087XE15 A0A3P9PGV7 A0A3Q1IID3 Q7SYR1 A0A1W4WZ88 A0A3B3U9G9 A0A146XJ08 A0A3B3R4S0 A0A1S3N1L8 A0A147AIE0 A0A3Q3T2J8 A0A1J1J4I1 A0A3Q2EIG5 A0A3B5R332 Q0V9I9 G3MH93 A0A0N7ZDA9
A0A336L7E3 A0A336L8H4 A0A1L8DTJ2 D6WGD4 A0A1B6LLM1 N6URV1 A0A1B6IXS4 U4UYN0 A0A1B6FX76 Q17JQ2 A0A1Q3F2H3 Q17JQ4 A0A182G8M0 A0A1S4F082 J3JXV9 B0WRY1 A0A1B6E6R6 A0A182N996 A0A1Y1L519 A0A182TT82 Q7QIM5 A0A2J7QBB4 A0A182VIC0 A0A182L3L9 A0A182I6S2 A0A182WWJ1 E0VCY3 A0A131XE09 W8C880 A0A1I8JUP0 A0A182QUX9 A0A3B3DUR2 A0A2M4BST6 A0A0K8R5M8 A0A182IR96 A0A182Y2C8 C4WVI6 A0A2P8XPS1 A0A2M4BST1 W5JPN9 A0A2M4ANZ2 A0A2M3Z9Y1 A0A3B4YWS2 A0A3Q4MCJ4 A0A3Q3JJ62 A0A2U9B1H6 A0A3P8TVH2 A0A182SXW4 A0A182PII1 I3K266 A0A1S3JUE1 A0A3Q3JCH8 A0A182JP77 Q5RHZ6 A0A3P8QCK5 A0A3B4H8Y6 A0A3Q3BRA2 A0A3P9C9B6 Q4SY05 A0A3B4UVE1 A0A131Z5L4 A0A067R3I0 A0A3B4YJJ8 A0A034VQT7 A0A224YIZ2 L7MF93 A0A1L8I071 A0A3Q2XSL8 A0A0S7MDV1 A0A3P9AJ18 A0A2D0T2K0 E3TBP5 A0A1E1XQA4 A0A0K8V745 A0A146N6M5 A0A232FHF2 A0A154PDK5 A0A087XE15 A0A3P9PGV7 A0A3Q1IID3 Q7SYR1 A0A1W4WZ88 A0A3B3U9G9 A0A146XJ08 A0A3B3R4S0 A0A1S3N1L8 A0A147AIE0 A0A3Q3T2J8 A0A1J1J4I1 A0A3Q2EIG5 A0A3B5R332 Q0V9I9 G3MH93 A0A0N7ZDA9
PDB
2VWB
E-value=3.31405e-90,
Score=845
Ontologies
PANTHER
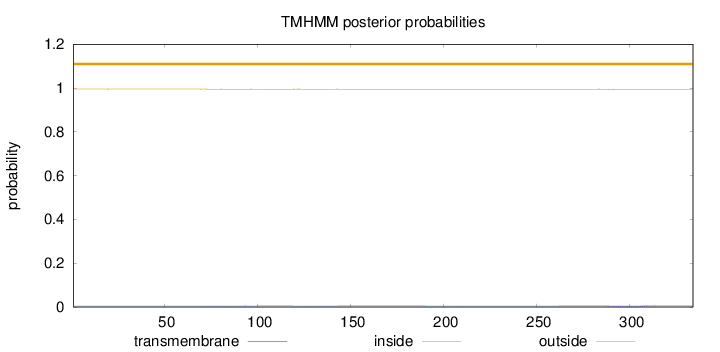
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
334
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08563
Exp number, first 60 AAs:
0.00048
Total prob of N-in:
0.00472
outside
1 - 334
Population Genetic Test Statistics
Pi
15.210799
Theta
12.848247
Tajima's D
-0.53261
CLR
2.734817
CSRT
0.233338333083346
Interpretation
Uncertain
