Gene
KWMTBOMO06782
Pre Gene Modal
BGIBMGA012036
Annotation
PREDICTED:_uncharacterized_protein_LOC100286771_isoform_X1_[Bombyx_mori]
Full name
Mini-chromosome maintenance complex-binding protein
Location in the cell
Nuclear Reliability : 3.222
Sequence
CDS
ATGCATGAAAATATCATGAGCGATATCGAAACAACGACGCCGGATCGATGGATTGAGAATGAAAAAATAATGAAGAGCAAGTTGATGAAATTACCAAATTGGTACCAGATCCCACTAATTAATTCAAATGCTTCACATAATTTAACAGACGGAAACTTAGTGCGGTTCAGAGGCATGATCCAAGACATGCACAACCCTGAATTTTACTTTGAGAAATTTGAAGTATTTAACACAACAACAAATGAAGTAAAGGTTAAAAGTGGAAAATACAGAGATACTGCACATGTTTTGGAAAATGAGAAAATTAACTATTCTGAGAACCTAATCAGTGGTCAGCGACAGACTTTAGTTGTTGTGTCTATACCTGGGTTAAATGACTGGGTCCAACAACTAGAAGATAAACAAAACTATTTGAAACATTTAGAAGAACCACCAAATACTTCCAAAAGGCTTCACCCAAGTACAAAGTTAAAAAGAAGCTACGATGATACAGAAGATAATTTGGATGAAATGGAAGTTGATAATGAAGCTGTGAATAAGGAAATGAAGATTTTAGATCACAAAGATAATACACCTGATGTGGTTTCAAGAGAACATTTGTTAAATTATCCTCTACCTGATGTACCTAGCAAATCTTGCATTGTTAAGATTTACAGTGACAATGAAAATCTCAAGCTCAATGACATGATAGAAGTCATTGGATTTCTTTCTGTAGATCCATCCCTATCAGGCGAATTCAAGGAAGATAAGGATTCTATGATAGAGCTCCAGATTGAGAGTAGTGTCGAAATTATAACTCATAATCCTCCACCAAGCTTAGTACCACGATTGCATGCTGTTCATGTACAAAAATTGGAACATTGCAATCCATTACTTTCAAATAGTGTTGATCAAGGTGTGATTTTAAGTGAGGCAAACGCAGCAAGGGAGCATTTATTGAAAGCCTTGACGGAACTCTTACTGGGAGATCAATTAGCTGCTGAATATTTGATATGTCATCTTATTGCTTGTGTCTATCTTCGTCAAGATACTATGAACTTGGGACAGTTCTGTTTGAACCTATCAAATTTGCCAACACAGAAATATCCAAATTATGCACAACAGCTATATGATATTATCAAGCAATTTGTCACCAAAAGCTACTATTTACCACTTACCATTGAAAATATGAATACATTGACATTATTGCCTAAGAAAGACTATGAATGTAATCGTCTAGCTAGTGGAGTACTACAGCTCAGTAAAGATACGCATCTAATTTTGGATGAAACCAAGATGGAACAAGGTCGGCTCGATGCGACCGGCGTCAACAATATAGCTGCTCTCGGTAAAATGATTATGACACAGAAAGTTGAATATGATTTCAAATATTATAAAATGGAATTTGATTCAGATATCTCTGTGCTGGTATTGTCTGAAGGAAAGTCATTGTTGCCAAGTGATTACCATGTTCCATTGAAACCAGAAGAATCTTCACTAGAAATATTTGATGCCATAGTTGAAGCAGCCACATACTACTTAACGGAGGACTTGATGAATATGATCAGGGCGTATCTAACCAACCTGAAACTTGTGAAATATTCTATAACTGAAGACTTGCAGTTTGTAGAAAATGATTTCATAGATATGAGAAGTAAATCAAGCAGTGATAATCCCGTAACGGCAGACGATTTACATAGATTACTGGTGTTGGCGCGCCTGGTCAGTTTATCGCGCGGCCACGATACTCTAAGTAAAGAATGCTGGGACATCACAAAGGCAATGGAAACTGAAAGATTACATCGAGTGAAGAATAGAGTTGCCTCTACTGTATAA
Protein
MHENIMSDIETTTPDRWIENEKIMKSKLMKLPNWYQIPLINSNASHNLTDGNLVRFRGMIQDMHNPEFYFEKFEVFNTTTNEVKVKSGKYRDTAHVLENEKINYSENLISGQRQTLVVVSIPGLNDWVQQLEDKQNYLKHLEEPPNTSKRLHPSTKLKRSYDDTEDNLDEMEVDNEAVNKEMKILDHKDNTPDVVSREHLLNYPLPDVPSKSCIVKIYSDNENLKLNDMIEVIGFLSVDPSLSGEFKEDKDSMIELQIESSVEIITHNPPPSLVPRLHAVHVQKLEHCNPLLSNSVDQGVILSEANAAREHLLKALTELLLGDQLAAEYLICHLIACVYLRQDTMNLGQFCLNLSNLPTQKYPNYAQQLYDIIKQFVTKSYYLPLTIENMNTLTLLPKKDYECNRLASGVLQLSKDTHLILDETKMEQGRLDATGVNNIAALGKMIMTQKVEYDFKYYKMEFDSDISVLVLSEGKSLLPSDYHVPLKPEESSLEIFDAIVEAATYYLTEDLMNMIRAYLTNLKLVKYSITEDLQFVENDFIDMRSKSSSDNPVTADDLHRLLVLARLVSLSRGHDTLSKECWDITKAMETERLHRVKNRVASTV
Summary
Description
Associated component of the MCM complex that acts as a regulator of DNA replication. Binds to the MCM complex during late S phase and may act by promoting the disassembly of the MCM complex from chromatin (By similarity).
Associated component of the MCM complex that acts as a regulator of DNA replication. Binds to the MCM complex during late S phase and promotes the disassembly of the MCM complex from chromatin, thereby acting as a key regulator of pre-replication complex (pre-RC) unloading from replicated DNA. Can dissociate the mcm complex without addition of ATP; probably acts by destabilizing interactions of each individual subunits of the MCM complex. May also be required for sister chromatid cohesion.
Associated component of the MCM complex that acts as a regulator of DNA replication. Binds to the MCM complex during late S phase and promotes the disassembly of the MCM complex from chromatin, thereby acting as a key regulator of pre-replication complex (pre-RC) unloading from replicated DNA. Can dissociate the mcm complex without addition of ATP; probably acts by destabilizing interactions of each individual subunits of the MCM complex. May also be required for sister chromatid cohesion.
Subunit
Interacts with the MCM complex.
Interacts with the mcm complex: associates with the mcm3-7 complex which lacks mcm2, while it does not interact with the mcm complex when mcm2 is present (mcm2-7 complex). Interacts with mcm7.
Interacts with the mcm complex: associates with the mcm3-7 complex which lacks mcm2, while it does not interact with the mcm complex when mcm2 is present (mcm2-7 complex). Interacts with mcm7.
Similarity
Belongs to the MCMBP family.
Keywords
Cell cycle
Cell division
Complete proteome
DNA replication
Mitosis
Nucleus
Phosphoprotein
Reference proteome
Feature
chain Mini-chromosome maintenance complex-binding protein
Uniprot
H9JR76
A0A2H1V9Y4
A0A2A4J3Z2
A0A1E1WQ04
A0A194Q9P4
A0A194QPQ7
+ More
A0A212F7Z6 S4PCJ5 D6WEZ4 A0A2J7PVP2 A0A1W4XKV9 A0A0T6B6P9 Q0IG57 A0A1S4F502 U4UHS8 W5JVF0 B4KJV3 N6TYT0 A0A182IP45 A0A182GT23 A0A067R5U4 A0A1I8N5Q3 B4LRG5 A0A1I8PYV3 A0A336LJY3 A0A3B0JGR3 A0A182FLN1 B4N062 Q29JX3 B4GL02 A0A1W4XLQ7 B4JEE6 A0A1W4UQI8 A0A1S3J8J2 A0A182QD81 A0A182PU86 W6JGW1 A0A151K3B2 A0A0L0CPP5 E2C2X0 A0A1A7X043 B4HY07 B4Q5A4 B3MKS6 B4NZY4 A0A1A8QX33 A0A182W2F8 A0A1A8LKD8 A0A1A8VHZ4 Q9VM60 A0A1B0F940 A0A1A8JLG5 A0A182RQZ9 A0A1B0C7Y4 A0A1A8BJY4 A0A182HNC4 A0A154PRE9 A0A182K614 A0A182N9L8 A0A182XC26 A0A1A8HF75 A0A182M851 A0A3B4A2Z9 A0A182VEA4 A0A1A9VIR3 A0A182UKI8 A0A151X3Q5 A0A1B0A6D8 Q7QGH1 F4WB70 A0A084VRL5 A0A0A1WKU0 A0A3Q4HJB6 A0A1A8FG05 A0A1B0GAR3 B3N671 A0A088ANV9 A0A195CN43 A0A1A9X0J6 K7J9J8 R7UQG6 A0A3P9BYS6 A0A0K8V9G6 A0A3B4YZA7 A0A0M4E937 A0A3P9N1N7 U5EXQ7 A0A3P8QMP1 A0A3Q2VEP8 A0A3B4F2V8 A0A3B4E151 Q7ZYP6 A0A3B4CDT0 A0A3P8UBQ0 A0A3Q1AUK7 A0A2I4D5F4 A0A3B3TZC2 F6XH58 A0A3Q2YM48
A0A212F7Z6 S4PCJ5 D6WEZ4 A0A2J7PVP2 A0A1W4XKV9 A0A0T6B6P9 Q0IG57 A0A1S4F502 U4UHS8 W5JVF0 B4KJV3 N6TYT0 A0A182IP45 A0A182GT23 A0A067R5U4 A0A1I8N5Q3 B4LRG5 A0A1I8PYV3 A0A336LJY3 A0A3B0JGR3 A0A182FLN1 B4N062 Q29JX3 B4GL02 A0A1W4XLQ7 B4JEE6 A0A1W4UQI8 A0A1S3J8J2 A0A182QD81 A0A182PU86 W6JGW1 A0A151K3B2 A0A0L0CPP5 E2C2X0 A0A1A7X043 B4HY07 B4Q5A4 B3MKS6 B4NZY4 A0A1A8QX33 A0A182W2F8 A0A1A8LKD8 A0A1A8VHZ4 Q9VM60 A0A1B0F940 A0A1A8JLG5 A0A182RQZ9 A0A1B0C7Y4 A0A1A8BJY4 A0A182HNC4 A0A154PRE9 A0A182K614 A0A182N9L8 A0A182XC26 A0A1A8HF75 A0A182M851 A0A3B4A2Z9 A0A182VEA4 A0A1A9VIR3 A0A182UKI8 A0A151X3Q5 A0A1B0A6D8 Q7QGH1 F4WB70 A0A084VRL5 A0A0A1WKU0 A0A3Q4HJB6 A0A1A8FG05 A0A1B0GAR3 B3N671 A0A088ANV9 A0A195CN43 A0A1A9X0J6 K7J9J8 R7UQG6 A0A3P9BYS6 A0A0K8V9G6 A0A3B4YZA7 A0A0M4E937 A0A3P9N1N7 U5EXQ7 A0A3P8QMP1 A0A3Q2VEP8 A0A3B4F2V8 A0A3B4E151 Q7ZYP6 A0A3B4CDT0 A0A3P8UBQ0 A0A3Q1AUK7 A0A2I4D5F4 A0A3B3TZC2 F6XH58 A0A3Q2YM48
Pubmed
19121390
26354079
22118469
23622113
18362917
19820115
+ More
17510324 23537049 20920257 23761445 17994087 26483478 24845553 25315136 15632085 24567405 26108605 20798317 22936249 17550304 10731132 12537572 12537569 18327897 25463417 12364791 21719571 24438588 25830018 25186727 20075255 23254933 21196493 20431018
17510324 23537049 20920257 23761445 17994087 26483478 24845553 25315136 15632085 24567405 26108605 20798317 22936249 17550304 10731132 12537572 12537569 18327897 25463417 12364791 21719571 24438588 25830018 25186727 20075255 23254933 21196493 20431018
EMBL
BABH01016366
ODYU01001443
SOQ37648.1
NWSH01003235
PCG66711.1
GDQN01001921
+ More
JAT89133.1 KQ459252 KPJ02129.1 KQ461185 KPJ07487.1 AGBW02009812 OWR49862.1 GAIX01004171 JAA88389.1 KQ971319 EFA00312.1 NEVH01020944 PNF20404.1 LJIG01009595 KRT82765.1 CH477274 EAT45154.1 KB632169 ERL89430.1 ADMH02000225 ETN67308.1 CH933807 EDW12556.1 APGK01049519 KB741156 ENN73511.1 JXUM01086088 KQ563543 KXJ73685.1 KK852867 KDR14717.1 CH940649 EDW64635.1 UFQT01000039 SSX18504.1 OUUW01000006 SPP81547.1 CH963920 EDW77997.1 CH379062 EAL32838.1 CH479184 EDW37318.1 CH916368 EDW03666.1 AXCN02002036 AB734108 BAO48218.1 LKEY01056763 KYN50624.1 JRES01000080 KNC34348.1 GL452255 EFN77674.1 HADW01009918 SBP11318.1 CH480818 EDW51937.1 CM000361 CM002910 EDX04037.1 KMY88662.1 CH902620 EDV31607.1 CM000157 EDW87811.1 HAEH01013804 HAEI01016947 SBR98360.1 HAEF01006982 HAEF01016192 HAEG01009914 SBR44364.1 HAEJ01018730 SBS59187.1 AE014134 AY069585 HAEE01000144 SBR20160.1 JXJN01025925 HADZ01003563 HAEA01014057 SBP67504.1 APCN01001904 KQ435007 KZC13690.1 HAEC01013911 SBQ82128.1 AXCM01003888 KQ982557 KYQ55043.1 AAAB01008834 EAA05693.4 GL888057 EGI68577.1 ATLV01015746 KE525033 KFB40609.1 GBXI01014638 JAC99653.1 HAEB01010537 SBQ57064.1 CCAG010015502 CH954177 EDV59158.1 KQ977532 KYN02128.1 AAZX01002692 AMQN01006769 KB299094 ELU08435.1 GDHF01017109 JAI35205.1 CP012523 ALC38638.1 GANO01000827 JAB59044.1 BC042336 AAMC01103138 AAMC01103139
JAT89133.1 KQ459252 KPJ02129.1 KQ461185 KPJ07487.1 AGBW02009812 OWR49862.1 GAIX01004171 JAA88389.1 KQ971319 EFA00312.1 NEVH01020944 PNF20404.1 LJIG01009595 KRT82765.1 CH477274 EAT45154.1 KB632169 ERL89430.1 ADMH02000225 ETN67308.1 CH933807 EDW12556.1 APGK01049519 KB741156 ENN73511.1 JXUM01086088 KQ563543 KXJ73685.1 KK852867 KDR14717.1 CH940649 EDW64635.1 UFQT01000039 SSX18504.1 OUUW01000006 SPP81547.1 CH963920 EDW77997.1 CH379062 EAL32838.1 CH479184 EDW37318.1 CH916368 EDW03666.1 AXCN02002036 AB734108 BAO48218.1 LKEY01056763 KYN50624.1 JRES01000080 KNC34348.1 GL452255 EFN77674.1 HADW01009918 SBP11318.1 CH480818 EDW51937.1 CM000361 CM002910 EDX04037.1 KMY88662.1 CH902620 EDV31607.1 CM000157 EDW87811.1 HAEH01013804 HAEI01016947 SBR98360.1 HAEF01006982 HAEF01016192 HAEG01009914 SBR44364.1 HAEJ01018730 SBS59187.1 AE014134 AY069585 HAEE01000144 SBR20160.1 JXJN01025925 HADZ01003563 HAEA01014057 SBP67504.1 APCN01001904 KQ435007 KZC13690.1 HAEC01013911 SBQ82128.1 AXCM01003888 KQ982557 KYQ55043.1 AAAB01008834 EAA05693.4 GL888057 EGI68577.1 ATLV01015746 KE525033 KFB40609.1 GBXI01014638 JAC99653.1 HAEB01010537 SBQ57064.1 CCAG010015502 CH954177 EDV59158.1 KQ977532 KYN02128.1 AAZX01002692 AMQN01006769 KB299094 ELU08435.1 GDHF01017109 JAI35205.1 CP012523 ALC38638.1 GANO01000827 JAB59044.1 BC042336 AAMC01103138 AAMC01103139
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000235965 UP000192223 UP000008820 UP000030742 UP000000673 UP000009192 UP000019118 UP000075880 UP000069940 UP000249989 UP000027135 UP000095301 UP000008792 UP000095300 UP000268350 UP000069272 UP000007798 UP000001819 UP000008744 UP000001070 UP000192221 UP000085678 UP000075886 UP000075885 UP000078492 UP000037069 UP000008237 UP000001292 UP000000304 UP000007801 UP000002282 UP000075920 UP000000803 UP000092443 UP000075900 UP000092460 UP000075840 UP000076502 UP000075881 UP000075884 UP000076407 UP000075883 UP000261520 UP000075903 UP000078200 UP000075902 UP000075809 UP000092445 UP000007062 UP000007755 UP000030765 UP000261580 UP000092444 UP000008711 UP000005203 UP000078542 UP000091820 UP000002358 UP000014760 UP000265160 UP000261400 UP000092553 UP000242638 UP000265100 UP000264840 UP000261460 UP000261440 UP000265080 UP000257160 UP000192220 UP000261500 UP000008143 UP000264820
UP000235965 UP000192223 UP000008820 UP000030742 UP000000673 UP000009192 UP000019118 UP000075880 UP000069940 UP000249989 UP000027135 UP000095301 UP000008792 UP000095300 UP000268350 UP000069272 UP000007798 UP000001819 UP000008744 UP000001070 UP000192221 UP000085678 UP000075886 UP000075885 UP000078492 UP000037069 UP000008237 UP000001292 UP000000304 UP000007801 UP000002282 UP000075920 UP000000803 UP000092443 UP000075900 UP000092460 UP000075840 UP000076502 UP000075881 UP000075884 UP000076407 UP000075883 UP000261520 UP000075903 UP000078200 UP000075902 UP000075809 UP000092445 UP000007062 UP000007755 UP000030765 UP000261580 UP000092444 UP000008711 UP000005203 UP000078542 UP000091820 UP000002358 UP000014760 UP000265160 UP000261400 UP000092553 UP000242638 UP000265100 UP000264840 UP000261460 UP000261440 UP000265080 UP000257160 UP000192220 UP000261500 UP000008143 UP000264820
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JR76
A0A2H1V9Y4
A0A2A4J3Z2
A0A1E1WQ04
A0A194Q9P4
A0A194QPQ7
+ More
A0A212F7Z6 S4PCJ5 D6WEZ4 A0A2J7PVP2 A0A1W4XKV9 A0A0T6B6P9 Q0IG57 A0A1S4F502 U4UHS8 W5JVF0 B4KJV3 N6TYT0 A0A182IP45 A0A182GT23 A0A067R5U4 A0A1I8N5Q3 B4LRG5 A0A1I8PYV3 A0A336LJY3 A0A3B0JGR3 A0A182FLN1 B4N062 Q29JX3 B4GL02 A0A1W4XLQ7 B4JEE6 A0A1W4UQI8 A0A1S3J8J2 A0A182QD81 A0A182PU86 W6JGW1 A0A151K3B2 A0A0L0CPP5 E2C2X0 A0A1A7X043 B4HY07 B4Q5A4 B3MKS6 B4NZY4 A0A1A8QX33 A0A182W2F8 A0A1A8LKD8 A0A1A8VHZ4 Q9VM60 A0A1B0F940 A0A1A8JLG5 A0A182RQZ9 A0A1B0C7Y4 A0A1A8BJY4 A0A182HNC4 A0A154PRE9 A0A182K614 A0A182N9L8 A0A182XC26 A0A1A8HF75 A0A182M851 A0A3B4A2Z9 A0A182VEA4 A0A1A9VIR3 A0A182UKI8 A0A151X3Q5 A0A1B0A6D8 Q7QGH1 F4WB70 A0A084VRL5 A0A0A1WKU0 A0A3Q4HJB6 A0A1A8FG05 A0A1B0GAR3 B3N671 A0A088ANV9 A0A195CN43 A0A1A9X0J6 K7J9J8 R7UQG6 A0A3P9BYS6 A0A0K8V9G6 A0A3B4YZA7 A0A0M4E937 A0A3P9N1N7 U5EXQ7 A0A3P8QMP1 A0A3Q2VEP8 A0A3B4F2V8 A0A3B4E151 Q7ZYP6 A0A3B4CDT0 A0A3P8UBQ0 A0A3Q1AUK7 A0A2I4D5F4 A0A3B3TZC2 F6XH58 A0A3Q2YM48
A0A212F7Z6 S4PCJ5 D6WEZ4 A0A2J7PVP2 A0A1W4XKV9 A0A0T6B6P9 Q0IG57 A0A1S4F502 U4UHS8 W5JVF0 B4KJV3 N6TYT0 A0A182IP45 A0A182GT23 A0A067R5U4 A0A1I8N5Q3 B4LRG5 A0A1I8PYV3 A0A336LJY3 A0A3B0JGR3 A0A182FLN1 B4N062 Q29JX3 B4GL02 A0A1W4XLQ7 B4JEE6 A0A1W4UQI8 A0A1S3J8J2 A0A182QD81 A0A182PU86 W6JGW1 A0A151K3B2 A0A0L0CPP5 E2C2X0 A0A1A7X043 B4HY07 B4Q5A4 B3MKS6 B4NZY4 A0A1A8QX33 A0A182W2F8 A0A1A8LKD8 A0A1A8VHZ4 Q9VM60 A0A1B0F940 A0A1A8JLG5 A0A182RQZ9 A0A1B0C7Y4 A0A1A8BJY4 A0A182HNC4 A0A154PRE9 A0A182K614 A0A182N9L8 A0A182XC26 A0A1A8HF75 A0A182M851 A0A3B4A2Z9 A0A182VEA4 A0A1A9VIR3 A0A182UKI8 A0A151X3Q5 A0A1B0A6D8 Q7QGH1 F4WB70 A0A084VRL5 A0A0A1WKU0 A0A3Q4HJB6 A0A1A8FG05 A0A1B0GAR3 B3N671 A0A088ANV9 A0A195CN43 A0A1A9X0J6 K7J9J8 R7UQG6 A0A3P9BYS6 A0A0K8V9G6 A0A3B4YZA7 A0A0M4E937 A0A3P9N1N7 U5EXQ7 A0A3P8QMP1 A0A3Q2VEP8 A0A3B4F2V8 A0A3B4E151 Q7ZYP6 A0A3B4CDT0 A0A3P8UBQ0 A0A3Q1AUK7 A0A2I4D5F4 A0A3B3TZC2 F6XH58 A0A3Q2YM48
Ontologies
GO
PANTHER
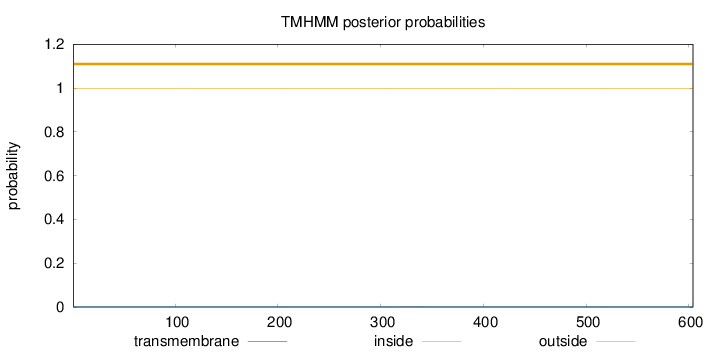
Topology
Subcellular location
Nucleus
Length:
604
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01279
Exp number, first 60 AAs:
0.00159
Total prob of N-in:
0.00077
outside
1 - 604
Population Genetic Test Statistics
Pi
19.863347
Theta
18.031825
Tajima's D
0.330104
CLR
13.734361
CSRT
0.479426028698565
Interpretation
Uncertain
