Pre Gene Modal
BGIBMGA012119
Annotation
PREDICTED:_integrin-linked_protein_kinase_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.462
Sequence
CDS
ATGTCGTGGGGCTCGCTGCACGCGCTGCTGCACGGCGGCGCGGGCGGGCGCGTGGTGGTGGACGCGGCGGCGGCGCTGCGGCTGGCGCACGACGTGGCGCGCGGCATGCGCTACCTGCACTCGCTGCAGCGGGACACCATCCTGCCCGCCTACCATCTCAACAGCAAACACATCATGATCGACGAAGACCTGACGGCCCGCATCAACATGGCGGACGCGAAGTTTTCGTTCCAAGAGCGCGGGCGGGTGTACGCGCCGGCGTGGATGGCCCCCGAGGCTCTGCTCAAGCCGGCCGCCAAGCGGAACTGGGAGGCCGCAGACATGTGGAGCTTCGCCGTGCTGTTGTGGGAGCTGGCCACCAGAGAGATACCATTCGCGGATCTGTCACCAATGGAGTGCGGCATGAAGATAGCGCTTGAAGGTCTTCGTATAAGCATACCTCCGGGCGTGTCGCCTCACATCTCGAAACTCATCAAGATCTGCATGAACGAGGATCCCGGGAAACGACCATCTTTCGAAATGATCCTACCGATTTTAGAGAAGATGAAACGCTGA
Protein
MSWGSLHALLHGGAGGRVVVDAAAALRLAHDVARGMRYLHSLQRDTILPAYHLNSKHIMIDEDLTARINMADAKFSFQERGRVYAPAWMAPEALLKPAAKRNWEAADMWSFAVLLWELATREIPFADLSPMECGMKIALEGLRISIPPGVSPHISKLIKICMNEDPGKRPSFEMILPILEKMKR
Summary
Uniprot
H9JRF9
A0A194QF86
A0A2A4J4B2
A0A2H1V9X2
Q2PQN4
A0A067QZH3
+ More
A0A2P8YW21 A0A2J7QNW1 T1PBM4 A0A1B0DFS8 A0A084WMQ5 A0A1A9WRB6 A0A023EQP6 A0A0T6BET7 A0A182QMF2 A0A1I8NVD5 A0A0L0CQN7 D3TMU2 A0A0V0G632 A0A182K9X6 A0A182PDI6 A0A182WBT2 A0A0K8TL72 A0A182ISR3 A0A182NIT3 A0A182Y2M7 A0A182I4F6 A0A182UM62 A0A182X4F6 A0A182LKH4 Q7Q595 A0A182UGR4 A0A1B0CHI2 A0A336N3S5 A0A0M4EL95 Q17PJ5 A0A182GXR3 Q2LYQ3 B4H5Y4 A0A3B0KB23 A0A069DU47 A0A224XPM3 A0A1L8DMB4 A0A1L8DMM9 A0A0P4W1V4 T1HQS0 A0A1W4V9C4 D6WHQ1 W8BI94 A0A182MH03 B3M6S7 W5J4S2 A0A023F5N9 A0A1W4WJJ0 A0A0A9Z7V8 A0A2M4AP43 B4IAL3 B3NIX2 B4QJP7 B4ITH9 A0A2M4ANP2 A0A1Q3F7G1 A0A1B6ED08 A0A336L995 C6ZQT0 B0XAZ0 A0A1Y1K652 A0A0L7LLV2 Q9V400 E0W1Q9 A0A0A1X3W0 B4LHA7 A0A1B0FBN4 A0A1A9VYN9 A0A1A9Z3A6 A0A1A9XWL4 A0A1B0B581 A0A2P6K757 U5EZ52 A0A023ESJ3 A0A2P6K074 B4KUL7 A0A154PJN0 A0A0K8WLI1 A0A158NB51 B4J133 A0A151J4D7 A0A195EVS0 B4ML54 A0A034VUS5 A0A0M8ZXY8 A0A0L7R987 A0A088A9C2 F4WHC0 E1ZYB0 A0A1B6LU99 A0A0J7KR97 A0A026W1E6 E2BM02 A0A195D6K8
A0A2P8YW21 A0A2J7QNW1 T1PBM4 A0A1B0DFS8 A0A084WMQ5 A0A1A9WRB6 A0A023EQP6 A0A0T6BET7 A0A182QMF2 A0A1I8NVD5 A0A0L0CQN7 D3TMU2 A0A0V0G632 A0A182K9X6 A0A182PDI6 A0A182WBT2 A0A0K8TL72 A0A182ISR3 A0A182NIT3 A0A182Y2M7 A0A182I4F6 A0A182UM62 A0A182X4F6 A0A182LKH4 Q7Q595 A0A182UGR4 A0A1B0CHI2 A0A336N3S5 A0A0M4EL95 Q17PJ5 A0A182GXR3 Q2LYQ3 B4H5Y4 A0A3B0KB23 A0A069DU47 A0A224XPM3 A0A1L8DMB4 A0A1L8DMM9 A0A0P4W1V4 T1HQS0 A0A1W4V9C4 D6WHQ1 W8BI94 A0A182MH03 B3M6S7 W5J4S2 A0A023F5N9 A0A1W4WJJ0 A0A0A9Z7V8 A0A2M4AP43 B4IAL3 B3NIX2 B4QJP7 B4ITH9 A0A2M4ANP2 A0A1Q3F7G1 A0A1B6ED08 A0A336L995 C6ZQT0 B0XAZ0 A0A1Y1K652 A0A0L7LLV2 Q9V400 E0W1Q9 A0A0A1X3W0 B4LHA7 A0A1B0FBN4 A0A1A9VYN9 A0A1A9Z3A6 A0A1A9XWL4 A0A1B0B581 A0A2P6K757 U5EZ52 A0A023ESJ3 A0A2P6K074 B4KUL7 A0A154PJN0 A0A0K8WLI1 A0A158NB51 B4J133 A0A151J4D7 A0A195EVS0 B4ML54 A0A034VUS5 A0A0M8ZXY8 A0A0L7R987 A0A088A9C2 F4WHC0 E1ZYB0 A0A1B6LU99 A0A0J7KR97 A0A026W1E6 E2BM02 A0A195D6K8
Pubmed
19121390
26354079
16907828
24845553
29403074
25315136
+ More
24438588 24945155 26108605 20353571 26369729 25244985 20966253 12364791 14747013 17210077 17510324 26483478 15632085 17994087 26334808 27129103 18362917 19820115 24495485 20920257 23761445 25474469 25401762 26823975 22936249 17550304 20496585 28004739 26227816 10637513 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 25830018 21347285 18057021 25348373 21719571 20798317 24508170
24438588 24945155 26108605 20353571 26369729 25244985 20966253 12364791 14747013 17210077 17510324 26483478 15632085 17994087 26334808 27129103 18362917 19820115 24495485 20920257 23761445 25474469 25401762 26823975 22936249 17550304 20496585 28004739 26227816 10637513 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 25830018 21347285 18057021 25348373 21719571 20798317 24508170
EMBL
BABH01016368
KQ459252
KPJ02131.1
NWSH01003235
PCG66709.1
ODYU01001443
+ More
SOQ37650.1 DQ307189 ABC25089.1 KK853074 KDR11771.1 PYGN01000327 PSN48447.1 NEVH01012562 PNF30233.1 KA646166 AFP60795.1 AJVK01058967 ATLV01024501 KE525352 KFB51499.1 GAPW01002178 JAC11420.1 LJIG01001233 KRT85707.1 AXCN02000480 JRES01000036 KNC34690.1 EZ422744 ADD19020.1 GECL01002604 JAP03520.1 GDAI01002943 JAI14660.1 APCN01002307 AAAB01008960 EAA11332.2 AJWK01012393 UFQT01004167 SSX35547.1 CP012525 ALC44185.1 CH477191 EAT48622.1 JXUM01095997 JXUM01097423 KQ564341 KQ564236 KXJ72404.1 KXJ72544.1 CH379069 EAL29808.1 CH479212 EDW33201.1 OUUW01000009 SPP85340.1 GBGD01001449 JAC87440.1 GFTR01006313 JAW10113.1 GFDF01006485 JAV07599.1 GFDF01006484 JAV07600.1 GDKW01000975 JAI55620.1 ACPB03023230 KQ971331 EEZ99715.1 GAMC01009852 GAMC01009851 JAB96703.1 AXCM01005052 CH902618 EDV39763.1 ADMH02002078 ETN59472.1 GBBI01002227 JAC16485.1 GBHO01003613 GBRD01008500 GDHC01001625 JAG39991.1 JAG57321.1 JAQ17004.1 GGFK01009067 MBW42388.1 CH480826 EDW44326.1 CH954178 EDV52618.1 CM000363 CM002912 EDX11347.1 KMZ00952.1 CM000159 CH891699 EDW95073.1 EDW99692.1 GGFK01009078 MBW42399.1 GFDL01011602 JAV23443.1 GEDC01015660 GEDC01001521 JAS21638.1 JAS35777.1 UFQS01002629 UFQT01002629 SSX14396.1 SSX33806.1 EZ114881 ACU30934.1 DS232603 EDS43960.1 GEZM01091389 JAV56863.1 JTDY01000629 KOB76415.1 AE014296 AY051708 AJ249345 AAF51691.1 AAK93132.1 CAB77053.1 DS235873 EEB19641.1 GBXI01008852 JAD05440.1 CH940647 EDW70620.1 CCAG010001557 JXJN01008636 MWRG01022752 PRD22154.1 GANO01001419 JAB58452.1 GAPW01001371 JAC12227.1 MWRG01054555 PRD19721.1 CH933809 EDW18245.1 KQ434938 KZC12052.1 GDHF01000410 JAI51904.1 ADTU01010884 CH916366 CH916456 EDV96888.1 EDV98665.1 KQ980151 KYN17501.1 KQ981953 KYN32360.1 CH963847 EDW73112.1 GAKP01012773 JAC46179.1 KQ435828 KOX71849.1 KQ414627 KOC67432.1 GL888158 EGI66397.1 GL435204 EFN73809.1 GEBQ01012723 JAT27254.1 LBMM01004087 KMQ92821.1 KK107494 EZA49838.1 GL449100 EFN83251.1 KQ976760 KYN08530.1
SOQ37650.1 DQ307189 ABC25089.1 KK853074 KDR11771.1 PYGN01000327 PSN48447.1 NEVH01012562 PNF30233.1 KA646166 AFP60795.1 AJVK01058967 ATLV01024501 KE525352 KFB51499.1 GAPW01002178 JAC11420.1 LJIG01001233 KRT85707.1 AXCN02000480 JRES01000036 KNC34690.1 EZ422744 ADD19020.1 GECL01002604 JAP03520.1 GDAI01002943 JAI14660.1 APCN01002307 AAAB01008960 EAA11332.2 AJWK01012393 UFQT01004167 SSX35547.1 CP012525 ALC44185.1 CH477191 EAT48622.1 JXUM01095997 JXUM01097423 KQ564341 KQ564236 KXJ72404.1 KXJ72544.1 CH379069 EAL29808.1 CH479212 EDW33201.1 OUUW01000009 SPP85340.1 GBGD01001449 JAC87440.1 GFTR01006313 JAW10113.1 GFDF01006485 JAV07599.1 GFDF01006484 JAV07600.1 GDKW01000975 JAI55620.1 ACPB03023230 KQ971331 EEZ99715.1 GAMC01009852 GAMC01009851 JAB96703.1 AXCM01005052 CH902618 EDV39763.1 ADMH02002078 ETN59472.1 GBBI01002227 JAC16485.1 GBHO01003613 GBRD01008500 GDHC01001625 JAG39991.1 JAG57321.1 JAQ17004.1 GGFK01009067 MBW42388.1 CH480826 EDW44326.1 CH954178 EDV52618.1 CM000363 CM002912 EDX11347.1 KMZ00952.1 CM000159 CH891699 EDW95073.1 EDW99692.1 GGFK01009078 MBW42399.1 GFDL01011602 JAV23443.1 GEDC01015660 GEDC01001521 JAS21638.1 JAS35777.1 UFQS01002629 UFQT01002629 SSX14396.1 SSX33806.1 EZ114881 ACU30934.1 DS232603 EDS43960.1 GEZM01091389 JAV56863.1 JTDY01000629 KOB76415.1 AE014296 AY051708 AJ249345 AAF51691.1 AAK93132.1 CAB77053.1 DS235873 EEB19641.1 GBXI01008852 JAD05440.1 CH940647 EDW70620.1 CCAG010001557 JXJN01008636 MWRG01022752 PRD22154.1 GANO01001419 JAB58452.1 GAPW01001371 JAC12227.1 MWRG01054555 PRD19721.1 CH933809 EDW18245.1 KQ434938 KZC12052.1 GDHF01000410 JAI51904.1 ADTU01010884 CH916366 CH916456 EDV96888.1 EDV98665.1 KQ980151 KYN17501.1 KQ981953 KYN32360.1 CH963847 EDW73112.1 GAKP01012773 JAC46179.1 KQ435828 KOX71849.1 KQ414627 KOC67432.1 GL888158 EGI66397.1 GL435204 EFN73809.1 GEBQ01012723 JAT27254.1 LBMM01004087 KMQ92821.1 KK107494 EZA49838.1 GL449100 EFN83251.1 KQ976760 KYN08530.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000027135
UP000245037
UP000235965
+ More
UP000095301 UP000092462 UP000030765 UP000091820 UP000075886 UP000095300 UP000037069 UP000075881 UP000075885 UP000075920 UP000075880 UP000075884 UP000076408 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000075902 UP000092461 UP000092553 UP000008820 UP000069940 UP000249989 UP000001819 UP000008744 UP000268350 UP000015103 UP000192221 UP000007266 UP000075883 UP000007801 UP000000673 UP000192223 UP000001292 UP000008711 UP000000304 UP000002282 UP000002320 UP000037510 UP000000803 UP000009046 UP000008792 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000009192 UP000076502 UP000005205 UP000001070 UP000078492 UP000078541 UP000007798 UP000053105 UP000053825 UP000005203 UP000007755 UP000000311 UP000036403 UP000053097 UP000008237 UP000078542
UP000095301 UP000092462 UP000030765 UP000091820 UP000075886 UP000095300 UP000037069 UP000075881 UP000075885 UP000075920 UP000075880 UP000075884 UP000076408 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000075902 UP000092461 UP000092553 UP000008820 UP000069940 UP000249989 UP000001819 UP000008744 UP000268350 UP000015103 UP000192221 UP000007266 UP000075883 UP000007801 UP000000673 UP000192223 UP000001292 UP000008711 UP000000304 UP000002282 UP000002320 UP000037510 UP000000803 UP000009046 UP000008792 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000009192 UP000076502 UP000005205 UP000001070 UP000078492 UP000078541 UP000007798 UP000053105 UP000053825 UP000005203 UP000007755 UP000000311 UP000036403 UP000053097 UP000008237 UP000078542
Interpro
Gene 3D
ProteinModelPortal
H9JRF9
A0A194QF86
A0A2A4J4B2
A0A2H1V9X2
Q2PQN4
A0A067QZH3
+ More
A0A2P8YW21 A0A2J7QNW1 T1PBM4 A0A1B0DFS8 A0A084WMQ5 A0A1A9WRB6 A0A023EQP6 A0A0T6BET7 A0A182QMF2 A0A1I8NVD5 A0A0L0CQN7 D3TMU2 A0A0V0G632 A0A182K9X6 A0A182PDI6 A0A182WBT2 A0A0K8TL72 A0A182ISR3 A0A182NIT3 A0A182Y2M7 A0A182I4F6 A0A182UM62 A0A182X4F6 A0A182LKH4 Q7Q595 A0A182UGR4 A0A1B0CHI2 A0A336N3S5 A0A0M4EL95 Q17PJ5 A0A182GXR3 Q2LYQ3 B4H5Y4 A0A3B0KB23 A0A069DU47 A0A224XPM3 A0A1L8DMB4 A0A1L8DMM9 A0A0P4W1V4 T1HQS0 A0A1W4V9C4 D6WHQ1 W8BI94 A0A182MH03 B3M6S7 W5J4S2 A0A023F5N9 A0A1W4WJJ0 A0A0A9Z7V8 A0A2M4AP43 B4IAL3 B3NIX2 B4QJP7 B4ITH9 A0A2M4ANP2 A0A1Q3F7G1 A0A1B6ED08 A0A336L995 C6ZQT0 B0XAZ0 A0A1Y1K652 A0A0L7LLV2 Q9V400 E0W1Q9 A0A0A1X3W0 B4LHA7 A0A1B0FBN4 A0A1A9VYN9 A0A1A9Z3A6 A0A1A9XWL4 A0A1B0B581 A0A2P6K757 U5EZ52 A0A023ESJ3 A0A2P6K074 B4KUL7 A0A154PJN0 A0A0K8WLI1 A0A158NB51 B4J133 A0A151J4D7 A0A195EVS0 B4ML54 A0A034VUS5 A0A0M8ZXY8 A0A0L7R987 A0A088A9C2 F4WHC0 E1ZYB0 A0A1B6LU99 A0A0J7KR97 A0A026W1E6 E2BM02 A0A195D6K8
A0A2P8YW21 A0A2J7QNW1 T1PBM4 A0A1B0DFS8 A0A084WMQ5 A0A1A9WRB6 A0A023EQP6 A0A0T6BET7 A0A182QMF2 A0A1I8NVD5 A0A0L0CQN7 D3TMU2 A0A0V0G632 A0A182K9X6 A0A182PDI6 A0A182WBT2 A0A0K8TL72 A0A182ISR3 A0A182NIT3 A0A182Y2M7 A0A182I4F6 A0A182UM62 A0A182X4F6 A0A182LKH4 Q7Q595 A0A182UGR4 A0A1B0CHI2 A0A336N3S5 A0A0M4EL95 Q17PJ5 A0A182GXR3 Q2LYQ3 B4H5Y4 A0A3B0KB23 A0A069DU47 A0A224XPM3 A0A1L8DMB4 A0A1L8DMM9 A0A0P4W1V4 T1HQS0 A0A1W4V9C4 D6WHQ1 W8BI94 A0A182MH03 B3M6S7 W5J4S2 A0A023F5N9 A0A1W4WJJ0 A0A0A9Z7V8 A0A2M4AP43 B4IAL3 B3NIX2 B4QJP7 B4ITH9 A0A2M4ANP2 A0A1Q3F7G1 A0A1B6ED08 A0A336L995 C6ZQT0 B0XAZ0 A0A1Y1K652 A0A0L7LLV2 Q9V400 E0W1Q9 A0A0A1X3W0 B4LHA7 A0A1B0FBN4 A0A1A9VYN9 A0A1A9Z3A6 A0A1A9XWL4 A0A1B0B581 A0A2P6K757 U5EZ52 A0A023ESJ3 A0A2P6K074 B4KUL7 A0A154PJN0 A0A0K8WLI1 A0A158NB51 B4J133 A0A151J4D7 A0A195EVS0 B4ML54 A0A034VUS5 A0A0M8ZXY8 A0A0L7R987 A0A088A9C2 F4WHC0 E1ZYB0 A0A1B6LU99 A0A0J7KR97 A0A026W1E6 E2BM02 A0A195D6K8
PDB
3REP
E-value=7.95642e-56,
Score=545
Ontologies
KEGG
GO
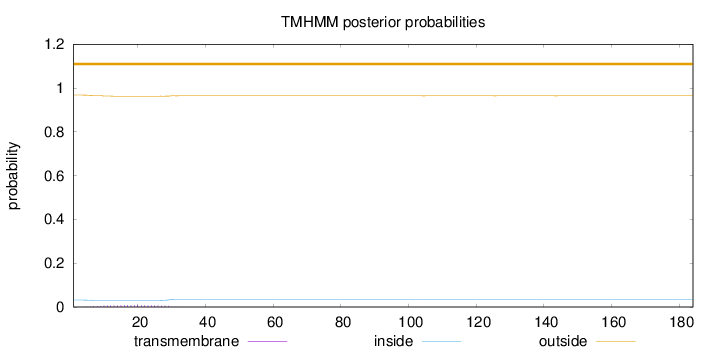
Topology
Length:
184
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17419
Exp number, first 60 AAs:
0.17193
Total prob of N-in:
0.03154
outside
1 - 184
Population Genetic Test Statistics
Pi
17.12464
Theta
18.055375
Tajima's D
-0.482173
CLR
0.423027
CSRT
0.24238788060597
Interpretation
Uncertain
