Gene
KWMTBOMO06777
Pre Gene Modal
BGIBMGA012035
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.824
Sequence
CDS
ATGGATTTAGAATTTCGAAATCTCACTTACACGGTGCAGAGTCGTATAGCTAAAGCAACAAAAAAGGTTGTCGTCAAAGGTGTCGACGGTAAATTCACGTCGGGCCAACTGGTCGCCATCATGGGGCCATCAGGAGCCGGCAAGAGTTCCCTTCTCAACATCATATCCGGTTATCGTTCAGCAGGAGTGACTGGCGAGCTCCTTGTGAACGGTCAGCCTCGAGACGAACACCAGTTCCAAAGATCGTCATGTTATATCACGCAAGAAGATCTATTGCAGCCACTTCTCACTGTCCGAGAGGCAATGGATGTCGCCGCTAAACTAAAACTACCCAGAGGTACCAAAGCTCCGTCCGAAGAAATACTCCAAGAGCTCGGACTCTTGGAGCATCAGAATACAAGGACCGATTCACTTTCGGGCGGACAGAAGAAAAGGTTATCGATTGCGCTAGAATTAGTTAACAACCCACCAGTATTCTTTCTCGACGAGCCAACTAGCGGACTAGATACAGTCACTACTACACAGTGTGTAAAGTTACTTAAGCAACTGGCTCGACAAGGAAGAACAATCGTGTGCACCATACATCAACCAGCGGCTTCGCTTCTCGAATTCTTTGATCAGCTCTATATAATAGCGGATGGATACTGTATATACCAAGGCGATACGGCAGCTATGGTGCCCTATCTTGAAAGTGTCGGTATGCCCTGTCCCAGACATCACAACCCTGCCGACTTCATCATCGAATTAACAGAGAACCCGGCTACTGTTAAACTATTGTCGCAAGAAATACTTAACGGAAAAATATACAAAACCTCCAAAATTGACCAGAACTGTACGGAACCGTTGCCACAGATGCAACTAAAGGTGTGCTATCAAGCCGAAGACAACGCACCAGAAACAGAAATTATGCTGACAAATGAAAAGTGTGTTTACAAAGTACAATCATACAGTAATACAGCTTCTATGTGTTCACTGAGTAGTCAGCTGTCCGGAATCAATAACTCCTCTGCATGGATGAAGATTCAAAAGCCTTCCGATTATGATATATCGTATTCCACTTCATATTTTGATCAATTTAGTATACTTCTCAAGAGGATGTTGCTGCAGATCTCAAGGAATAGGCAAGGGCTTTGGATCCAACTATTACATCACTTGATGTGCGGGCTTCTGATAGGAGCATGCTTCTATAATATGGCCAACGACGGAACAGAGATGTTTAACCACTTGAAGCTGTGTGTGGGAATTGTGATATTCTTCGCGTACACGCAAATCATGGTCCCAGTTTTAGTTTATCCACAAGAAGTTAAATTAGTGAAAAAAGAGTATTTTAACTGCTGGTACAGCTTGACTCCATACTACGCCGCGCTCACAGTCTCAAAGCTTCCAGTACAACTGTTTTGTAACATACTGTTCTCTACAATAGTTTACTTCATGGCTGGTATTCCATTCAACGTAATGAGGTTCATAACGTTCTGTGCCGTGGGGAACATGGTGTCTTTAGTCTCCGAAGGAATCGGCTTGGCTATAGGATCAGTCTTCAACGTCACGAACGGATGCGCAGTCGGTCCGTCAGCGATAGCGCCATTCCTGGGGCTAGGAGTCTACGGATTCGACTTTGCACACCAGATCCCGCTCATCATGAACATACTGATGAAGCTGTCGTTTATTCGATGTGGAATCGTCGCCATGGTCCTCACGGTGTTCGGGTTCGGAAGACAGCCTCTCGAGTGCAGCGAGGTTTTCTGTCATTTCGCTAAGCCAAACGTTCTAATGCAGTATTTAGACATCGACAAAAGCTCAGTCTGGATGGAAATCATGTTCCTCTTAATAATCATGTTAGTGTTCAGGTCGATGTGCTATATCAGTCTCAGGTGGAGGTTTGCGACGTGA
Protein
MDLEFRNLTYTVQSRIAKATKKVVVKGVDGKFTSGQLVAIMGPSGAGKSSLLNIISGYRSAGVTGELLVNGQPRDEHQFQRSSCYITQEDLLQPLLTVREAMDVAAKLKLPRGTKAPSEEILQELGLLEHQNTRTDSLSGGQKKRLSIALELVNNPPVFFLDEPTSGLDTVTTTQCVKLLKQLARQGRTIVCTIHQPAASLLEFFDQLYIIADGYCIYQGDTAAMVPYLESVGMPCPRHHNPADFIIELTENPATVKLLSQEILNGKIYKTSKIDQNCTEPLPQMQLKVCYQAEDNAPETEIMLTNEKCVYKVQSYSNTASMCSLSSQLSGINNSSAWMKIQKPSDYDISYSTSYFDQFSILLKRMLLQISRNRQGLWIQLLHHLMCGLLIGACFYNMANDGTEMFNHLKLCVGIVIFFAYTQIMVPVLVYPQEVKLVKKEYFNCWYSLTPYYAALTVSKLPVQLFCNILFSTIVYFMAGIPFNVMRFITFCAVGNMVSLVSEGIGLAIGSVFNVTNGCAVGPSAIAPFLGLGVYGFDFAHQIPLIMNILMKLSFIRCGIVAMVLTVFGFGRQPLECSEVFCHFAKPNVLMQYLDIDKSSVWMEIMFLLIIMLVFRSMCYISLRWRFAT
Summary
Uniprot
A0A3S2NR26
A0A223PQE0
A0A2H4KAY5
A0A2H1WHR2
A0A194PGA9
A0A212EPY7
+ More
A0A194R8X1 A0A0L7LFT1 A0A067R385 A0A2J7R9W6 E0VAP1 A0A1B6DAM8 A0A1B6DT64 A0A1B6J212 A0A0P4VY32 J9JSX9 T1I2C3 A0A2H8TW86 A0A336K6X9 A0A1Q3FNE6 A0A1Q3FNC5 A0A1B6JKA0 A0A0A9YIV2 U5ETU5 A0A1J1J8I3 A0A2M4BHD3 A0A182YC72 A0A182P1H7 A0A1Q3FNT7 A0A1L8E4C2 A0A3B0JVM2 A0A0J9RTT3 B4HEW3 B4KUR3 A0A1W4VU95 B4PFL8 Q2LZ69 Q9VTL3 Q8SXX6 B3NGY3 U5EW92 B4LCP8 A0A034V8F4 A0A0K8WFN7 A0A2H4TES0 A0A182K7V4 B4N3B8 A0A0A1WY89 A0A0M4EG47 W8AXI7 A0A1I8Q2H2 A0A1I8NFC6 A0A1B6HTY1 A0A182FC22 A0A067QKC2 Q16NT8 A0A2J7R9W9 A0A182WD55 W5JR55 Q8MS57 A0A182H727 A0A151WQQ6 A0A151J3U4 A0A2H8TD64 A0A195FT41 A0A2S2NCV5 A0A1L3IWL4 D1ZZE5 J9K4C5 A0A026WAG1 E2A629 A0A182RA65 A0A2S2Q1D6 A0A182G6U3 A0A1B0D4K3 A0A158V3H3 A0A1L8E4W0 A0A1W4X256 A0A0L0BM15 A0A2J7R9W2 A0A182YDK4 A0A182UY98 A0A2C9GPY6 F4W572 A0A2C9GQ51 A0A182X335 A0A1I8M5X7 Q7Q1C5 A0A182WGS4 E0W3T6 A0A1B6KJ17 A0A2H1WPU0 A0A0M4EDA3 E2C0H4 H9J6F2 A0A1B6KT77 A0A182JPJ5 A0A1I8PPX2
A0A194R8X1 A0A0L7LFT1 A0A067R385 A0A2J7R9W6 E0VAP1 A0A1B6DAM8 A0A1B6DT64 A0A1B6J212 A0A0P4VY32 J9JSX9 T1I2C3 A0A2H8TW86 A0A336K6X9 A0A1Q3FNE6 A0A1Q3FNC5 A0A1B6JKA0 A0A0A9YIV2 U5ETU5 A0A1J1J8I3 A0A2M4BHD3 A0A182YC72 A0A182P1H7 A0A1Q3FNT7 A0A1L8E4C2 A0A3B0JVM2 A0A0J9RTT3 B4HEW3 B4KUR3 A0A1W4VU95 B4PFL8 Q2LZ69 Q9VTL3 Q8SXX6 B3NGY3 U5EW92 B4LCP8 A0A034V8F4 A0A0K8WFN7 A0A2H4TES0 A0A182K7V4 B4N3B8 A0A0A1WY89 A0A0M4EG47 W8AXI7 A0A1I8Q2H2 A0A1I8NFC6 A0A1B6HTY1 A0A182FC22 A0A067QKC2 Q16NT8 A0A2J7R9W9 A0A182WD55 W5JR55 Q8MS57 A0A182H727 A0A151WQQ6 A0A151J3U4 A0A2H8TD64 A0A195FT41 A0A2S2NCV5 A0A1L3IWL4 D1ZZE5 J9K4C5 A0A026WAG1 E2A629 A0A182RA65 A0A2S2Q1D6 A0A182G6U3 A0A1B0D4K3 A0A158V3H3 A0A1L8E4W0 A0A1W4X256 A0A0L0BM15 A0A2J7R9W2 A0A182YDK4 A0A182UY98 A0A2C9GPY6 F4W572 A0A2C9GQ51 A0A182X335 A0A1I8M5X7 Q7Q1C5 A0A182WGS4 E0W3T6 A0A1B6KJ17 A0A2H1WPU0 A0A0M4EDA3 E2C0H4 H9J6F2 A0A1B6KT77 A0A182JPJ5 A0A1I8PPX2
Pubmed
26354079
22118469
26227816
24845553
20566863
27129103
+ More
25401762 26823975 25244985 22936249 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25348373 29121518 25830018 24495485 25315136 17510324 20920257 23761445 26483478 18362917 19820115 24508170 30249741 20798317 26108605 21719571 12364791 19121390
25401762 26823975 25244985 22936249 17994087 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25348373 29121518 25830018 24495485 25315136 17510324 20920257 23761445 26483478 18362917 19820115 24508170 30249741 20798317 26108605 21719571 12364791 19121390
EMBL
RSAL01000007
RVE54117.1
KX710205
ASU47349.1
KY484788
ASS36030.1
+ More
ODYU01008422 SOQ52004.1 KQ459606 KPI91739.1 AGBW02013361 OWR43521.1 KQ460779 KPJ12321.1 JTDY01001309 KOB74240.1 KK853032 KDR12258.1 NEVH01006580 PNF37625.1 DS235012 EEB10447.1 GEDC01029100 GEDC01026518 GEDC01025608 GEDC01014560 GEDC01010316 GEDC01006295 GEDC01001363 JAS08198.1 JAS10780.1 JAS11690.1 JAS22738.1 JAS26982.1 JAS31003.1 JAS35935.1 GEDC01030302 GEDC01030086 GEDC01008426 GEDC01005369 GEDC01000164 JAS06996.1 JAS07212.1 JAS28872.1 JAS31929.1 JAS37134.1 GECU01014556 GECU01002618 JAS93150.1 JAT05089.1 GDKW01001065 JAI55530.1 ABLF02034227 ABLF02034231 ABLF02034236 ACPB03006729 ACPB03006730 GFXV01006246 MBW18051.1 UFQS01000131 UFQT01000131 SSX00231.1 SSX20611.1 GFDL01006013 JAV29032.1 GFDL01006011 JAV29034.1 GECU01008086 JAS99620.1 GBHO01042572 GBHO01025113 GBHO01014144 GBHO01014141 GBHO01014139 GBRD01009136 GDHC01010334 GDHC01003672 JAG01032.1 JAG18491.1 JAG29460.1 JAG29463.1 JAG29465.1 JAG56685.1 JAQ08295.1 JAQ14957.1 GANO01002626 JAB57245.1 CVRI01000074 CRL08274.1 GGFJ01003262 MBW52403.1 GFDL01005899 JAV29146.1 GFDF01000511 JAV13573.1 OUUW01000002 SPP76781.1 CM002912 KMY99032.1 CH480815 EDW41132.1 CH933809 EDW19319.1 CM000159 EDW94167.2 CH379069 EAL29640.1 AE014296 BT056236 AAF50035.2 ACL68683.1 AY075521 AAL68329.1 CH954178 EDV51440.1 GANO01003054 JAB56817.1 CH940647 EDW70940.1 KRF85201.1 KRF85202.1 GAKP01020912 GAKP01020911 JAC38040.1 GDHF01002644 JAI49670.1 KY849643 ATY74524.1 CH964062 EDW78857.2 GBXI01010939 JAD03353.1 CP012525 ALC45046.1 GAMC01017047 JAB89508.1 GECU01029611 JAS78095.1 KK853336 KDR08336.1 CH477809 EAT36018.1 PNF37621.1 ADMH02000414 ETN66606.1 AY119083 AAM50943.1 JXUM01027850 JXUM01027851 JXUM01027852 KQ560803 KXJ80838.1 KQ982821 KYQ50170.1 KQ980249 KYN17094.1 GFXV01000246 MBW12051.1 KQ981276 KYN43611.1 GGMR01002391 MBY15010.1 KX056437 APG57147.1 KQ971338 EFA01863.2 ABLF02036674 KK107295 QOIP01000003 EZA53082.1 RLU24924.1 GL437108 EFN71088.1 GGMS01002375 MBY71578.1 JXUM01045625 JXUM01045626 JXUM01045627 KQ561445 KXJ78564.1 AJVK01003298 KF828798 AIN44115.1 GFDF01000512 JAV13572.1 JRES01001684 KNC20968.1 PNF37622.1 APCN01001726 GL887605 EGI70668.1 AAAB01008980 EAA14400.3 DS235883 EEB20292.1 GEBQ01028562 JAT11415.1 ODYU01009789 SOQ54444.1 CP012523 ALC39760.1 GL451783 EFN78567.1 BABH01019639 BABH01019640 GEBQ01025342 JAT14635.1
ODYU01008422 SOQ52004.1 KQ459606 KPI91739.1 AGBW02013361 OWR43521.1 KQ460779 KPJ12321.1 JTDY01001309 KOB74240.1 KK853032 KDR12258.1 NEVH01006580 PNF37625.1 DS235012 EEB10447.1 GEDC01029100 GEDC01026518 GEDC01025608 GEDC01014560 GEDC01010316 GEDC01006295 GEDC01001363 JAS08198.1 JAS10780.1 JAS11690.1 JAS22738.1 JAS26982.1 JAS31003.1 JAS35935.1 GEDC01030302 GEDC01030086 GEDC01008426 GEDC01005369 GEDC01000164 JAS06996.1 JAS07212.1 JAS28872.1 JAS31929.1 JAS37134.1 GECU01014556 GECU01002618 JAS93150.1 JAT05089.1 GDKW01001065 JAI55530.1 ABLF02034227 ABLF02034231 ABLF02034236 ACPB03006729 ACPB03006730 GFXV01006246 MBW18051.1 UFQS01000131 UFQT01000131 SSX00231.1 SSX20611.1 GFDL01006013 JAV29032.1 GFDL01006011 JAV29034.1 GECU01008086 JAS99620.1 GBHO01042572 GBHO01025113 GBHO01014144 GBHO01014141 GBHO01014139 GBRD01009136 GDHC01010334 GDHC01003672 JAG01032.1 JAG18491.1 JAG29460.1 JAG29463.1 JAG29465.1 JAG56685.1 JAQ08295.1 JAQ14957.1 GANO01002626 JAB57245.1 CVRI01000074 CRL08274.1 GGFJ01003262 MBW52403.1 GFDL01005899 JAV29146.1 GFDF01000511 JAV13573.1 OUUW01000002 SPP76781.1 CM002912 KMY99032.1 CH480815 EDW41132.1 CH933809 EDW19319.1 CM000159 EDW94167.2 CH379069 EAL29640.1 AE014296 BT056236 AAF50035.2 ACL68683.1 AY075521 AAL68329.1 CH954178 EDV51440.1 GANO01003054 JAB56817.1 CH940647 EDW70940.1 KRF85201.1 KRF85202.1 GAKP01020912 GAKP01020911 JAC38040.1 GDHF01002644 JAI49670.1 KY849643 ATY74524.1 CH964062 EDW78857.2 GBXI01010939 JAD03353.1 CP012525 ALC45046.1 GAMC01017047 JAB89508.1 GECU01029611 JAS78095.1 KK853336 KDR08336.1 CH477809 EAT36018.1 PNF37621.1 ADMH02000414 ETN66606.1 AY119083 AAM50943.1 JXUM01027850 JXUM01027851 JXUM01027852 KQ560803 KXJ80838.1 KQ982821 KYQ50170.1 KQ980249 KYN17094.1 GFXV01000246 MBW12051.1 KQ981276 KYN43611.1 GGMR01002391 MBY15010.1 KX056437 APG57147.1 KQ971338 EFA01863.2 ABLF02036674 KK107295 QOIP01000003 EZA53082.1 RLU24924.1 GL437108 EFN71088.1 GGMS01002375 MBY71578.1 JXUM01045625 JXUM01045626 JXUM01045627 KQ561445 KXJ78564.1 AJVK01003298 KF828798 AIN44115.1 GFDF01000512 JAV13572.1 JRES01001684 KNC20968.1 PNF37622.1 APCN01001726 GL887605 EGI70668.1 AAAB01008980 EAA14400.3 DS235883 EEB20292.1 GEBQ01028562 JAT11415.1 ODYU01009789 SOQ54444.1 CP012523 ALC39760.1 GL451783 EFN78567.1 BABH01019639 BABH01019640 GEBQ01025342 JAT14635.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000053240
UP000037510
UP000027135
+ More
UP000235965 UP000009046 UP000007819 UP000015103 UP000183832 UP000076408 UP000075885 UP000268350 UP000001292 UP000009192 UP000192221 UP000002282 UP000001819 UP000000803 UP000008711 UP000008792 UP000075881 UP000007798 UP000092553 UP000095300 UP000095301 UP000069272 UP000008820 UP000075920 UP000000673 UP000069940 UP000249989 UP000075809 UP000078492 UP000078541 UP000007266 UP000053097 UP000279307 UP000000311 UP000075900 UP000092462 UP000192223 UP000037069 UP000075903 UP000075840 UP000007755 UP000076407 UP000007062 UP000008237 UP000005204
UP000235965 UP000009046 UP000007819 UP000015103 UP000183832 UP000076408 UP000075885 UP000268350 UP000001292 UP000009192 UP000192221 UP000002282 UP000001819 UP000000803 UP000008711 UP000008792 UP000075881 UP000007798 UP000092553 UP000095300 UP000095301 UP000069272 UP000008820 UP000075920 UP000000673 UP000069940 UP000249989 UP000075809 UP000078492 UP000078541 UP000007266 UP000053097 UP000279307 UP000000311 UP000075900 UP000092462 UP000192223 UP000037069 UP000075903 UP000075840 UP000007755 UP000076407 UP000007062 UP000008237 UP000005204
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A3S2NR26
A0A223PQE0
A0A2H4KAY5
A0A2H1WHR2
A0A194PGA9
A0A212EPY7
+ More
A0A194R8X1 A0A0L7LFT1 A0A067R385 A0A2J7R9W6 E0VAP1 A0A1B6DAM8 A0A1B6DT64 A0A1B6J212 A0A0P4VY32 J9JSX9 T1I2C3 A0A2H8TW86 A0A336K6X9 A0A1Q3FNE6 A0A1Q3FNC5 A0A1B6JKA0 A0A0A9YIV2 U5ETU5 A0A1J1J8I3 A0A2M4BHD3 A0A182YC72 A0A182P1H7 A0A1Q3FNT7 A0A1L8E4C2 A0A3B0JVM2 A0A0J9RTT3 B4HEW3 B4KUR3 A0A1W4VU95 B4PFL8 Q2LZ69 Q9VTL3 Q8SXX6 B3NGY3 U5EW92 B4LCP8 A0A034V8F4 A0A0K8WFN7 A0A2H4TES0 A0A182K7V4 B4N3B8 A0A0A1WY89 A0A0M4EG47 W8AXI7 A0A1I8Q2H2 A0A1I8NFC6 A0A1B6HTY1 A0A182FC22 A0A067QKC2 Q16NT8 A0A2J7R9W9 A0A182WD55 W5JR55 Q8MS57 A0A182H727 A0A151WQQ6 A0A151J3U4 A0A2H8TD64 A0A195FT41 A0A2S2NCV5 A0A1L3IWL4 D1ZZE5 J9K4C5 A0A026WAG1 E2A629 A0A182RA65 A0A2S2Q1D6 A0A182G6U3 A0A1B0D4K3 A0A158V3H3 A0A1L8E4W0 A0A1W4X256 A0A0L0BM15 A0A2J7R9W2 A0A182YDK4 A0A182UY98 A0A2C9GPY6 F4W572 A0A2C9GQ51 A0A182X335 A0A1I8M5X7 Q7Q1C5 A0A182WGS4 E0W3T6 A0A1B6KJ17 A0A2H1WPU0 A0A0M4EDA3 E2C0H4 H9J6F2 A0A1B6KT77 A0A182JPJ5 A0A1I8PPX2
A0A194R8X1 A0A0L7LFT1 A0A067R385 A0A2J7R9W6 E0VAP1 A0A1B6DAM8 A0A1B6DT64 A0A1B6J212 A0A0P4VY32 J9JSX9 T1I2C3 A0A2H8TW86 A0A336K6X9 A0A1Q3FNE6 A0A1Q3FNC5 A0A1B6JKA0 A0A0A9YIV2 U5ETU5 A0A1J1J8I3 A0A2M4BHD3 A0A182YC72 A0A182P1H7 A0A1Q3FNT7 A0A1L8E4C2 A0A3B0JVM2 A0A0J9RTT3 B4HEW3 B4KUR3 A0A1W4VU95 B4PFL8 Q2LZ69 Q9VTL3 Q8SXX6 B3NGY3 U5EW92 B4LCP8 A0A034V8F4 A0A0K8WFN7 A0A2H4TES0 A0A182K7V4 B4N3B8 A0A0A1WY89 A0A0M4EG47 W8AXI7 A0A1I8Q2H2 A0A1I8NFC6 A0A1B6HTY1 A0A182FC22 A0A067QKC2 Q16NT8 A0A2J7R9W9 A0A182WD55 W5JR55 Q8MS57 A0A182H727 A0A151WQQ6 A0A151J3U4 A0A2H8TD64 A0A195FT41 A0A2S2NCV5 A0A1L3IWL4 D1ZZE5 J9K4C5 A0A026WAG1 E2A629 A0A182RA65 A0A2S2Q1D6 A0A182G6U3 A0A1B0D4K3 A0A158V3H3 A0A1L8E4W0 A0A1W4X256 A0A0L0BM15 A0A2J7R9W2 A0A182YDK4 A0A182UY98 A0A2C9GPY6 F4W572 A0A2C9GQ51 A0A182X335 A0A1I8M5X7 Q7Q1C5 A0A182WGS4 E0W3T6 A0A1B6KJ17 A0A2H1WPU0 A0A0M4EDA3 E2C0H4 H9J6F2 A0A1B6KT77 A0A182JPJ5 A0A1I8PPX2
PDB
6FFC
E-value=3.34366e-58,
Score=572
Ontologies
GO
Topology
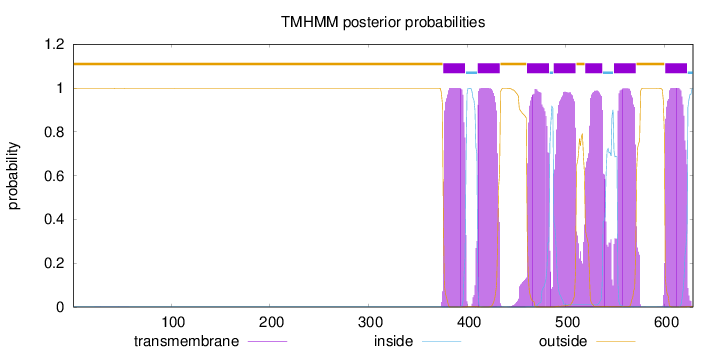
Length:
629
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
152.93753
Exp number, first 60 AAs:
0.01092
Total prob of N-in:
0.00159
outside
1 - 375
TMhelix
376 - 398
inside
399 - 410
TMhelix
411 - 433
outside
434 - 460
TMhelix
461 - 483
inside
484 - 487
TMhelix
488 - 510
outside
511 - 519
TMhelix
520 - 537
inside
538 - 548
TMhelix
549 - 571
outside
572 - 600
TMhelix
601 - 623
inside
624 - 629
Population Genetic Test Statistics
Pi
24.902941
Theta
23.503733
Tajima's D
0.559082
CLR
1.29227
CSRT
0.535573221338933
Interpretation
Uncertain
