Pre Gene Modal
BGIBMGA012120
Annotation
PREDICTED:_arf-GAP_with_dual_PH_domain-containing_protein_1-like_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.356
Sequence
CDS
ATGGTAGACCATAACCAGAAATTATTACAGGAGTTGCAAAAACGAAGTGGAAATAACGTGTGCGCCGACTGTTTAAATCCTGACCCAGACTGGGCTTCTTACAACCTTGGGATATTCATATGCATGCGATGTGCAAGCATCCACCGTGGCATGGGCGCTCATATCAGCAAAGTGAAACATTTGAAGCTGGACAGATGGGAAGACTCACAGGTACAGAGGATGAAGGAAGTCGGGAACGTAACTGCTAGGAATAAATACGAGGAACGAGTTCCCCCTTGCTACAGAAGACCTAATAGAAATGACCCTCAAGTCCTCATTGAACAATGGATCCGAGCCAAATATGAGCGCGAAGAATTCTGTCATCCCGAACGTCAGTCGTATTTGTCGGGAAGCATGCAGGGCTTCCTTATGAAACGAGGCAAGGAGGACAGCAGATACCAACTCAGAAAATTCATACTCAGTGAAACCGAAGATACGCTCAAGTATCACGTTAGAGAGGACAAGGAACCGAAGGGCGTTCTCAAATTGTCAGAGTTGAATGTTGCTTTCGCGCCACCAAAAATCGGCCACATGAACTCAATGCAGTTGACATTCATGAAAGATGGCACCACTAGGCACATCTATGTTTATCACGAGGAGCCTGAAGTAATAAGCAACTGGTATACAGCGATACGTTGTGCACAACTCCATCTACTTCAAGTTGCGTTTCCGTCGACTCCGGAGTCGGATCTCATTCATCGCCTACCAAGAGATTTCGCACGTGAAGGCTGGCTATGGAAGACCGGGCCCAGACCTACTGACGCTCATAGAAGACGCTGGTTCACCCTCGACAACAGAAAACTCATGTACCACGACGAGCCGCTTGATGCTTATCCCAAGGGCGAGATTTTTGTCGGTAACGAATCGAACGGCTACTGTATTAGAGTTTCAAATACGGGCAATGGCTGCAAGGAGGCGAGGTTCCCTTTCCAGCTGGTGACTCCAGAAAGAACGTACTGCCTCGCTGCGACAACACACGAGGACCGCGACTGTTGGCTAGCAGTTCTGCAGCAGACGATCAAAAGAAGTCTCACGCCTCAGGATTCTACCATTGAAGCTGTCCTTGTGAGGAAACGCAACACATCGAACCCGATCAACATTTTCTCGGGAAGGTAG
Protein
MVDHNQKLLQELQKRSGNNVCADCLNPDPDWASYNLGIFICMRCASIHRGMGAHISKVKHLKLDRWEDSQVQRMKEVGNVTARNKYEERVPPCYRRPNRNDPQVLIEQWIRAKYEREEFCHPERQSYLSGSMQGFLMKRGKEDSRYQLRKFILSETEDTLKYHVREDKEPKGVLKLSELNVAFAPPKIGHMNSMQLTFMKDGTTRHIYVYHEEPEVISNWYTAIRCAQLHLLQVAFPSTPESDLIHRLPRDFAREGWLWKTGPRPTDAHRRRWFTLDNRKLMYHDEPLDAYPKGEIFVGNESNGYCIRVSNTGNGCKEARFPFQLVTPERTYCLAATTHEDRDCWLAVLQQTIKRSLTPQDSTIEAVLVRKRNTSNPINIFSGR
Summary
Uniprot
A0A2A4JKR7
H9JRG0
A0A3S2M8T8
A0A2A4JK16
A0A194PEN7
A0A2H1VKP4
+ More
A0A1E1W9M1 A0A212EUC5 A0A1E1WD80 A0A194R471 A0A1L8DMM1 A0A158NHU9 A0A1W4XCU1 F4X3D2 A0A1Y1KAN3 A0A2A3E3E3 A0A088A3I4 A0A067QZI2 E2AVA7 A0A154NYS9 A0A0M9A6X2 K7IQS0 E2C8N7 A0A232FKG3 A0A0C9QS63 A0A195FFM7 A0A1Q3G0D3 A0A195AWG4 A0A195CDF8 A0A1L8DM36 A0A195EHB5 A0A151WQU8 A0A182K2A8 A0A182XYP9 A0A182JGZ2 A0A1Y9IW65 A0A182R7B5 A0A182PQL3 A0A182QCS4 A0A182M0I1 A0A2J7QUM6 A0A182UNU8 A0A084VD62 A0A182X3X3 A0A182KR47 Q7QJA2 Q0IEU3 A0A182UGB4 A0A0L7L9T4 A0A182I346 A0A3L8DIX3 A0A1S4FI57 A0A182NI20 A0A2M3Z8K3 A0A2M4AD41 A0A182FDE4 A0A2M4BR19 A0A0L7QS65 W5JFR0 A0A1Q3G0I2 D6WDB5 A0A0T6B811 A0A2M4ADF0 A0A2M4CYS1 A0A1W4XN58 T1HGT2 A0A026WU38 A0A1B6HMU7 A0A182HAX0 A0A1B6EEN5 A0A2R7WLC3 A0A1B6CL23 A0A0A9WIF1 A0A0P5HJZ1 V5I111 E9GXU5 A0A0P5M1F1 L7M2N1 A0A0N8ABT6 N6U9M3 B7Q985 C3Y8X2 A0A131X8Y9 A0A2R5LL84 A0A0P4Y363 A0A0P4WGN9 A0A1J1ICP4 A0A293LNA0 A0A0P4W8F4 C0JBT6 A0A1S3HND0 A0A1S3HNB0 A0A3R7PES1 B0XIK8 R7U8G8 A0A1E1X6G8 T1GG86 A0A3Q0IZ79 M3Z2H0 O02753
A0A1E1W9M1 A0A212EUC5 A0A1E1WD80 A0A194R471 A0A1L8DMM1 A0A158NHU9 A0A1W4XCU1 F4X3D2 A0A1Y1KAN3 A0A2A3E3E3 A0A088A3I4 A0A067QZI2 E2AVA7 A0A154NYS9 A0A0M9A6X2 K7IQS0 E2C8N7 A0A232FKG3 A0A0C9QS63 A0A195FFM7 A0A1Q3G0D3 A0A195AWG4 A0A195CDF8 A0A1L8DM36 A0A195EHB5 A0A151WQU8 A0A182K2A8 A0A182XYP9 A0A182JGZ2 A0A1Y9IW65 A0A182R7B5 A0A182PQL3 A0A182QCS4 A0A182M0I1 A0A2J7QUM6 A0A182UNU8 A0A084VD62 A0A182X3X3 A0A182KR47 Q7QJA2 Q0IEU3 A0A182UGB4 A0A0L7L9T4 A0A182I346 A0A3L8DIX3 A0A1S4FI57 A0A182NI20 A0A2M3Z8K3 A0A2M4AD41 A0A182FDE4 A0A2M4BR19 A0A0L7QS65 W5JFR0 A0A1Q3G0I2 D6WDB5 A0A0T6B811 A0A2M4ADF0 A0A2M4CYS1 A0A1W4XN58 T1HGT2 A0A026WU38 A0A1B6HMU7 A0A182HAX0 A0A1B6EEN5 A0A2R7WLC3 A0A1B6CL23 A0A0A9WIF1 A0A0P5HJZ1 V5I111 E9GXU5 A0A0P5M1F1 L7M2N1 A0A0N8ABT6 N6U9M3 B7Q985 C3Y8X2 A0A131X8Y9 A0A2R5LL84 A0A0P4Y363 A0A0P4WGN9 A0A1J1ICP4 A0A293LNA0 A0A0P4W8F4 C0JBT6 A0A1S3HND0 A0A1S3HNB0 A0A3R7PES1 B0XIK8 R7U8G8 A0A1E1X6G8 T1GG86 A0A3Q0IZ79 M3Z2H0 O02753
Pubmed
19121390
26354079
22118469
21347285
21719571
28004739
+ More
24845553 20798317 20075255 28648823 25244985 24438588 20966253 12364791 14747013 17210077 17510324 26227816 30249741 20920257 23761445 18362917 19820115 24508170 26483478 25401762 26823975 25765539 21292972 25576852 23537049 18563158 28049606 19073266 23254933 28503490 23236062 9151987
24845553 20798317 20075255 28648823 25244985 24438588 20966253 12364791 14747013 17210077 17510324 26227816 30249741 20920257 23761445 18362917 19820115 24508170 26483478 25401762 26823975 25765539 21292972 25576852 23537049 18563158 28049606 19073266 23254933 28503490 23236062 9151987
EMBL
NWSH01001146
PCG72409.1
BABH01016379
RSAL01000007
RVE54113.1
PCG72407.1
+ More
KQ459606 KPI91737.1 ODYU01003082 SOQ41393.1 GDQN01007415 JAT83639.1 AGBW02012418 OWR45088.1 GDQN01006145 JAT84909.1 KQ460779 KPJ12319.1 GFDF01006470 JAV07614.1 ADTU01016068 GL888609 EGI59113.1 GEZM01090663 JAV57210.1 KZ288403 PBC26218.1 KK852855 KDR14920.1 GL443059 EFN62661.1 KQ434783 KZC04722.1 KQ435724 KOX78485.1 GL453666 EFN75767.1 NNAY01000082 OXU31152.1 GBYB01003487 GBYB01009813 JAG73254.1 JAG79580.1 KQ981636 KYN38829.1 GFDL01001798 JAV33247.1 KQ976725 KYM76588.1 KQ978023 KYM98103.1 GFDF01006694 JAV07390.1 KQ978957 KYN27262.1 KQ982813 KYQ50282.1 AXCN02000387 AXCM01000608 NEVH01010503 PNF32280.1 ATLV01011185 KE524650 KFB35906.1 AAAB01008807 EAA04627.4 CH477478 EAT40258.1 JTDY01002041 KOB72258.1 APCN01000197 QOIP01000008 RLU19798.1 GGFM01004102 MBW24853.1 GGFK01005207 MBW38528.1 GGFJ01006369 MBW55510.1 KQ414758 KOC61473.1 ADMH02001280 ETN63212.1 GFDL01001753 JAV33292.1 KQ971321 EEZ99543.2 LJIG01009233 KRT83490.1 GGFK01005495 MBW38816.1 GGFL01006269 MBW70447.1 ACPB03006856 KK107109 EZA59141.1 GECU01031730 JAS75976.1 JXUM01031575 KQ560926 KXJ80322.1 GEDC01000905 JAS36393.1 KK855033 PTY20413.1 GEDC01023176 JAS14122.1 GBHO01036423 GBRD01002263 GDHC01020808 GDHC01003506 JAG07181.1 JAG63558.1 JAP97820.1 JAQ15123.1 GDIQ01225847 GDIP01112848 LRGB01002451 JAK25878.1 JAL90866.1 KZS07480.1 GANP01003510 JAB80958.1 GL732573 EFX75755.1 GDIQ01170965 JAK80760.1 GACK01006719 JAA58315.1 GDIP01162849 JAJ60553.1 APGK01034640 APGK01034641 APGK01034642 KB740914 KB632352 ENN78390.1 ERL93266.1 ABJB010033394 DS887784 EEC15407.1 GG666492 EEN63028.1 GEFH01005228 JAP63353.1 GGLE01006089 MBY10215.1 GDIP01233095 JAI90306.1 GDRN01064076 JAI64895.1 CVRI01000043 CRK96209.1 GFWV01003465 MAA28195.1 GDRN01064077 JAI64894.1 FJ172672 ACN32215.1 QCYY01002630 ROT68815.1 DS233328 EDS29432.1 AMQN01009828 KB306314 ELT99981.1 GFAC01004374 JAT94814.1 CAQQ02194750 CAQQ02194751 AEYP01088986 JP005159 AER93756.1 D89940 BAA20132.1
KQ459606 KPI91737.1 ODYU01003082 SOQ41393.1 GDQN01007415 JAT83639.1 AGBW02012418 OWR45088.1 GDQN01006145 JAT84909.1 KQ460779 KPJ12319.1 GFDF01006470 JAV07614.1 ADTU01016068 GL888609 EGI59113.1 GEZM01090663 JAV57210.1 KZ288403 PBC26218.1 KK852855 KDR14920.1 GL443059 EFN62661.1 KQ434783 KZC04722.1 KQ435724 KOX78485.1 GL453666 EFN75767.1 NNAY01000082 OXU31152.1 GBYB01003487 GBYB01009813 JAG73254.1 JAG79580.1 KQ981636 KYN38829.1 GFDL01001798 JAV33247.1 KQ976725 KYM76588.1 KQ978023 KYM98103.1 GFDF01006694 JAV07390.1 KQ978957 KYN27262.1 KQ982813 KYQ50282.1 AXCN02000387 AXCM01000608 NEVH01010503 PNF32280.1 ATLV01011185 KE524650 KFB35906.1 AAAB01008807 EAA04627.4 CH477478 EAT40258.1 JTDY01002041 KOB72258.1 APCN01000197 QOIP01000008 RLU19798.1 GGFM01004102 MBW24853.1 GGFK01005207 MBW38528.1 GGFJ01006369 MBW55510.1 KQ414758 KOC61473.1 ADMH02001280 ETN63212.1 GFDL01001753 JAV33292.1 KQ971321 EEZ99543.2 LJIG01009233 KRT83490.1 GGFK01005495 MBW38816.1 GGFL01006269 MBW70447.1 ACPB03006856 KK107109 EZA59141.1 GECU01031730 JAS75976.1 JXUM01031575 KQ560926 KXJ80322.1 GEDC01000905 JAS36393.1 KK855033 PTY20413.1 GEDC01023176 JAS14122.1 GBHO01036423 GBRD01002263 GDHC01020808 GDHC01003506 JAG07181.1 JAG63558.1 JAP97820.1 JAQ15123.1 GDIQ01225847 GDIP01112848 LRGB01002451 JAK25878.1 JAL90866.1 KZS07480.1 GANP01003510 JAB80958.1 GL732573 EFX75755.1 GDIQ01170965 JAK80760.1 GACK01006719 JAA58315.1 GDIP01162849 JAJ60553.1 APGK01034640 APGK01034641 APGK01034642 KB740914 KB632352 ENN78390.1 ERL93266.1 ABJB010033394 DS887784 EEC15407.1 GG666492 EEN63028.1 GEFH01005228 JAP63353.1 GGLE01006089 MBY10215.1 GDIP01233095 JAI90306.1 GDRN01064076 JAI64895.1 CVRI01000043 CRK96209.1 GFWV01003465 MAA28195.1 GDRN01064077 JAI64894.1 FJ172672 ACN32215.1 QCYY01002630 ROT68815.1 DS233328 EDS29432.1 AMQN01009828 KB306314 ELT99981.1 GFAC01004374 JAT94814.1 CAQQ02194750 CAQQ02194751 AEYP01088986 JP005159 AER93756.1 D89940 BAA20132.1
Proteomes
UP000218220
UP000005204
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000005205 UP000192223 UP000007755 UP000242457 UP000005203 UP000027135 UP000000311 UP000076502 UP000053105 UP000002358 UP000008237 UP000215335 UP000078541 UP000078540 UP000078542 UP000078492 UP000075809 UP000075881 UP000076408 UP000075880 UP000075920 UP000075900 UP000075885 UP000075886 UP000075883 UP000235965 UP000075903 UP000030765 UP000076407 UP000075882 UP000007062 UP000008820 UP000075902 UP000037510 UP000075840 UP000279307 UP000075884 UP000069272 UP000053825 UP000000673 UP000007266 UP000015103 UP000053097 UP000069940 UP000249989 UP000076858 UP000000305 UP000019118 UP000030742 UP000001555 UP000001554 UP000183832 UP000085678 UP000283509 UP000002320 UP000014760 UP000015102 UP000079169 UP000000715
UP000005205 UP000192223 UP000007755 UP000242457 UP000005203 UP000027135 UP000000311 UP000076502 UP000053105 UP000002358 UP000008237 UP000215335 UP000078541 UP000078540 UP000078542 UP000078492 UP000075809 UP000075881 UP000076408 UP000075880 UP000075920 UP000075900 UP000075885 UP000075886 UP000075883 UP000235965 UP000075903 UP000030765 UP000076407 UP000075882 UP000007062 UP000008820 UP000075902 UP000037510 UP000075840 UP000279307 UP000075884 UP000069272 UP000053825 UP000000673 UP000007266 UP000015103 UP000053097 UP000069940 UP000249989 UP000076858 UP000000305 UP000019118 UP000030742 UP000001555 UP000001554 UP000183832 UP000085678 UP000283509 UP000002320 UP000014760 UP000015102 UP000079169 UP000000715
PRIDE
Pfam
Interpro
IPR001849
PH_domain
+ More
IPR011993 PH-like_dom_sf
IPR037278 ARFGAP/RecO
IPR037849 PH1_ADAP
IPR037851 PH2_ADAP
IPR001164 ArfGAP_dom
IPR038508 ArfGAP_dom_sf
IPR005725 ATPase_V1-cplx_asu
IPR004100 ATPase_F1/V1/A1_a/bsu_N
IPR023366 ATP_synth_asu-like_sf
IPR031686 ATP-synth_a_Xtn
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR024034 ATPase_F1/V1_b/a_C
IPR022878 V-ATPase_asu
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR020003 ATPase_a/bsu_AS
IPR027417 P-loop_NTPase
IPR011993 PH-like_dom_sf
IPR037278 ARFGAP/RecO
IPR037849 PH1_ADAP
IPR037851 PH2_ADAP
IPR001164 ArfGAP_dom
IPR038508 ArfGAP_dom_sf
IPR005725 ATPase_V1-cplx_asu
IPR004100 ATPase_F1/V1/A1_a/bsu_N
IPR023366 ATP_synth_asu-like_sf
IPR031686 ATP-synth_a_Xtn
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR024034 ATPase_F1/V1_b/a_C
IPR022878 V-ATPase_asu
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR020003 ATPase_a/bsu_AS
IPR027417 P-loop_NTPase
Gene 3D
ProteinModelPortal
A0A2A4JKR7
H9JRG0
A0A3S2M8T8
A0A2A4JK16
A0A194PEN7
A0A2H1VKP4
+ More
A0A1E1W9M1 A0A212EUC5 A0A1E1WD80 A0A194R471 A0A1L8DMM1 A0A158NHU9 A0A1W4XCU1 F4X3D2 A0A1Y1KAN3 A0A2A3E3E3 A0A088A3I4 A0A067QZI2 E2AVA7 A0A154NYS9 A0A0M9A6X2 K7IQS0 E2C8N7 A0A232FKG3 A0A0C9QS63 A0A195FFM7 A0A1Q3G0D3 A0A195AWG4 A0A195CDF8 A0A1L8DM36 A0A195EHB5 A0A151WQU8 A0A182K2A8 A0A182XYP9 A0A182JGZ2 A0A1Y9IW65 A0A182R7B5 A0A182PQL3 A0A182QCS4 A0A182M0I1 A0A2J7QUM6 A0A182UNU8 A0A084VD62 A0A182X3X3 A0A182KR47 Q7QJA2 Q0IEU3 A0A182UGB4 A0A0L7L9T4 A0A182I346 A0A3L8DIX3 A0A1S4FI57 A0A182NI20 A0A2M3Z8K3 A0A2M4AD41 A0A182FDE4 A0A2M4BR19 A0A0L7QS65 W5JFR0 A0A1Q3G0I2 D6WDB5 A0A0T6B811 A0A2M4ADF0 A0A2M4CYS1 A0A1W4XN58 T1HGT2 A0A026WU38 A0A1B6HMU7 A0A182HAX0 A0A1B6EEN5 A0A2R7WLC3 A0A1B6CL23 A0A0A9WIF1 A0A0P5HJZ1 V5I111 E9GXU5 A0A0P5M1F1 L7M2N1 A0A0N8ABT6 N6U9M3 B7Q985 C3Y8X2 A0A131X8Y9 A0A2R5LL84 A0A0P4Y363 A0A0P4WGN9 A0A1J1ICP4 A0A293LNA0 A0A0P4W8F4 C0JBT6 A0A1S3HND0 A0A1S3HNB0 A0A3R7PES1 B0XIK8 R7U8G8 A0A1E1X6G8 T1GG86 A0A3Q0IZ79 M3Z2H0 O02753
A0A1E1W9M1 A0A212EUC5 A0A1E1WD80 A0A194R471 A0A1L8DMM1 A0A158NHU9 A0A1W4XCU1 F4X3D2 A0A1Y1KAN3 A0A2A3E3E3 A0A088A3I4 A0A067QZI2 E2AVA7 A0A154NYS9 A0A0M9A6X2 K7IQS0 E2C8N7 A0A232FKG3 A0A0C9QS63 A0A195FFM7 A0A1Q3G0D3 A0A195AWG4 A0A195CDF8 A0A1L8DM36 A0A195EHB5 A0A151WQU8 A0A182K2A8 A0A182XYP9 A0A182JGZ2 A0A1Y9IW65 A0A182R7B5 A0A182PQL3 A0A182QCS4 A0A182M0I1 A0A2J7QUM6 A0A182UNU8 A0A084VD62 A0A182X3X3 A0A182KR47 Q7QJA2 Q0IEU3 A0A182UGB4 A0A0L7L9T4 A0A182I346 A0A3L8DIX3 A0A1S4FI57 A0A182NI20 A0A2M3Z8K3 A0A2M4AD41 A0A182FDE4 A0A2M4BR19 A0A0L7QS65 W5JFR0 A0A1Q3G0I2 D6WDB5 A0A0T6B811 A0A2M4ADF0 A0A2M4CYS1 A0A1W4XN58 T1HGT2 A0A026WU38 A0A1B6HMU7 A0A182HAX0 A0A1B6EEN5 A0A2R7WLC3 A0A1B6CL23 A0A0A9WIF1 A0A0P5HJZ1 V5I111 E9GXU5 A0A0P5M1F1 L7M2N1 A0A0N8ABT6 N6U9M3 B7Q985 C3Y8X2 A0A131X8Y9 A0A2R5LL84 A0A0P4Y363 A0A0P4WGN9 A0A1J1ICP4 A0A293LNA0 A0A0P4W8F4 C0JBT6 A0A1S3HND0 A0A1S3HNB0 A0A3R7PES1 B0XIK8 R7U8G8 A0A1E1X6G8 T1GG86 A0A3Q0IZ79 M3Z2H0 O02753
PDB
3MDB
E-value=1.36271e-98,
Score=918
Ontologies
GO
PANTHER
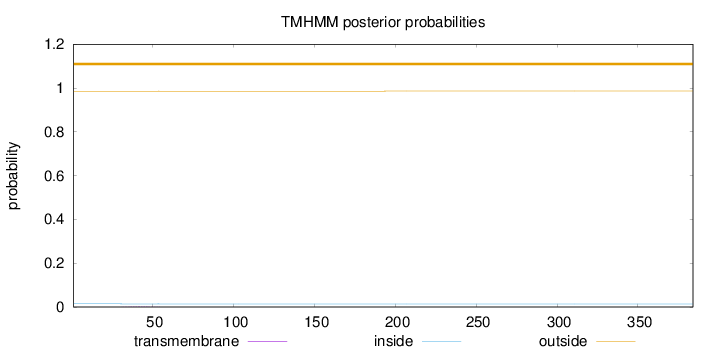
Topology
Length:
384
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02759
Exp number, first 60 AAs:
0.02744
Total prob of N-in:
0.01454
outside
1 - 384
Population Genetic Test Statistics
Pi
265.293939
Theta
185.888319
Tajima's D
1.161737
CLR
0.000364
CSRT
0.701514924253787
Interpretation
Uncertain
