Gene
KWMTBOMO06769 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012123
Annotation
Pyridoxal-5'-phosphate-dependent_protein_beta_subunit_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.958
Sequence
CDS
ATGGCAAACAATAATAGCGTTGACAACGAAATCAAGGCATCAGCCTTGGATCTCATCGGCAATACGCCTATAGTAGCGCTCGATCGCCTTCACCCTGGCCCCGGAAGGATCTTGGCGAAATGTGAGTTCATGAACCCGGGAGCATCTATTAAGTGCCGGTCATCACTTTGCATGATCAAGAAAGCGTTGGCCTCTGGTGAACTGAAACCTAAAGAGCCAGTTGTGGAAGTGACATCGGGAAATCAAGGATGTGGGCTCGCCGTCGTGTGTGCTGTATTGGGTCATCCGCTAACGCTTGCCATGTCAAAAGGAAACAGTGCTCAAAGGGCCATACATATGGAGGCTCTCGGGGCCAAATGCGTAAGAGTTCCGCAAGTGGAAGGCACGTACGGAAACGTGACTTTGAACGATGTAAAAGCAGTGGAGGAAGTAGGCCTAAGAATTGTAAAGGAAACCGGCGCATATTATGTGAATCAGTTCAACAACGACGCCAACTCCGAGTCTCACTACCAAACCACAGGACCTGAGATATGGAGACAGACTGGACATCGTGTTGACGCATTTGTTGCTACTGTCGGTACCGCGGGCACTTTTACTGGAACTACAAGGTATTTAAAGGAAAAGAATCCAGATTTGAAGGCGTACGTTGTTGAACCTGAAGGCGCTCAACCAATAAGAGGACTGCCAGTCATAAAGCCCCTTCATCTCCTACAAGGGTCAGGTTACGGTTGCGTTCCGAATTTATTCAAACGTGAGTACATGGATGGCACCCTGAGCGTATCTGATGAGGAAGCGATGGAGTATAAAAAGCTGATTGGCGAGAAAGAAGGTTTATATGTGGGCTACACAAGTGCTGCTAATGTAGCAGCGGCCGTCAAGTTGCTGAAGTCTGGACAGTTGCCTGAGGATTCGTGGGTAGTCACCATGCTCAATGACACCGGCTTGAAATATACTCCAGTGCCTGAAGAGTTTACGTAA
Protein
MANNNSVDNEIKASALDLIGNTPIVALDRLHPGPGRILAKCEFMNPGASIKCRSSLCMIKKALASGELKPKEPVVEVTSGNQGCGLAVVCAVLGHPLTLAMSKGNSAQRAIHMEALGAKCVRVPQVEGTYGNVTLNDVKAVEEVGLRIVKETGAYYVNQFNNDANSESHYQTTGPEIWRQTGHRVDAFVATVGTAGTFTGTTRYLKEKNPDLKAYVVEPEGAQPIRGLPVIKPLHLLQGSGYGCVPNLFKREYMDGTLSVSDEEAMEYKKLIGEKEGLYVGYTSAANVAAAVKLLKSGQLPEDSWVVTMLNDTGLKYTPVPEEFT
Summary
Uniprot
H9JRG3
A0A0L7LB36
A0A0U3S5Y4
A0A220K8H3
A0A220K819
A0A194PFD1
+ More
A0A2H1VR62 A0A194R8W1 A0A2A4J5Y7 A0A220K8C9 A0A220K835 A0A220K8J3 A0A2A4J4L8 A0A220K8A8 A0A220K813 A0A2H1W7D9 A0A220K8E3 A0A220K8D1 A0A220K8F1 A0A0U3QZ45 A0A220K8B9 I4DLE5 A0A212EQW4 A0A220K8D8 A0A220K824 A0A1D2MWT8 A0A0U3SCD8 A0A212FH38 A0A220K8D2 T1KF23 A0A023ZRZ4 A0A226DGG8 A0A3S3NRW7 A0A0L6J4R0 A0A1I3RB50 A0A1I4IDD8 I9WTW2 A0A0V7Z7W8 A0A0S8G065 A0A3G2V492 A0A1I7FYA1 A0A0E9KEZ5 B1M078 A0A2M8VFB9 A0A1I6PQ87 A0A2U8X9Z7 A0A2U1E2Q1 A0A1F9AHE9 A0A2A4B330 A0A0J6T7G5 A3JXC5 A0A354Y7X9 A0A225MD82 W1KQD9 A0A2M8QIC4 A0A239IFX7 T0GU46 A0A2T4HNU1 A0A1T5ESV9 A0A126RG36 T0I0H1 A0A1T5GLM8 A0A031IXS0 A0A126RF03 A0A081SXB7 A0A1E1F8E0 A0A2K8WBZ3 A0A0M7MJC7 A0A1R1K2W7 A0A2I8DC77 A0A1I6VCR4 A0A365XMT5 A0A0M7MMQ9 A0A0D6IXP8 A0A2M9GRQ0 A0A1D8IJF5 A0A0X8P154 A0A0M7K288 A0A0M7HQQ6 A0A179T477 A1HQX6 F6DQI8 F7SZI9 A0A0B5DW83 A0A1S6IVG3 A0A1M6TM50 A0A1F8QFU6 A0A1M4VVD8 A4J492 A0A1G0YYH4 K8DXV2 A0A2C6ME27 A0A3N5KT09 W7S7N8 A0A3P3G6S4 A0A2A3DNU8 A0A0A0DZP8 A0A2R6CDD4 A0A1F8RW04 A0A3S2YAM0
A0A2H1VR62 A0A194R8W1 A0A2A4J5Y7 A0A220K8C9 A0A220K835 A0A220K8J3 A0A2A4J4L8 A0A220K8A8 A0A220K813 A0A2H1W7D9 A0A220K8E3 A0A220K8D1 A0A220K8F1 A0A0U3QZ45 A0A220K8B9 I4DLE5 A0A212EQW4 A0A220K8D8 A0A220K824 A0A1D2MWT8 A0A0U3SCD8 A0A212FH38 A0A220K8D2 T1KF23 A0A023ZRZ4 A0A226DGG8 A0A3S3NRW7 A0A0L6J4R0 A0A1I3RB50 A0A1I4IDD8 I9WTW2 A0A0V7Z7W8 A0A0S8G065 A0A3G2V492 A0A1I7FYA1 A0A0E9KEZ5 B1M078 A0A2M8VFB9 A0A1I6PQ87 A0A2U8X9Z7 A0A2U1E2Q1 A0A1F9AHE9 A0A2A4B330 A0A0J6T7G5 A3JXC5 A0A354Y7X9 A0A225MD82 W1KQD9 A0A2M8QIC4 A0A239IFX7 T0GU46 A0A2T4HNU1 A0A1T5ESV9 A0A126RG36 T0I0H1 A0A1T5GLM8 A0A031IXS0 A0A126RF03 A0A081SXB7 A0A1E1F8E0 A0A2K8WBZ3 A0A0M7MJC7 A0A1R1K2W7 A0A2I8DC77 A0A1I6VCR4 A0A365XMT5 A0A0M7MMQ9 A0A0D6IXP8 A0A2M9GRQ0 A0A1D8IJF5 A0A0X8P154 A0A0M7K288 A0A0M7HQQ6 A0A179T477 A1HQX6 F6DQI8 F7SZI9 A0A0B5DW83 A0A1S6IVG3 A0A1M6TM50 A0A1F8QFU6 A0A1M4VVD8 A4J492 A0A1G0YYH4 K8DXV2 A0A2C6ME27 A0A3N5KT09 W7S7N8 A0A3P3G6S4 A0A2A3DNU8 A0A0A0DZP8 A0A2R6CDD4 A0A1F8RW04 A0A3S2YAM0
Pubmed
EMBL
BABH01016384
JTDY01001966
KOB72436.1
KT358809
ALV66640.1
MF038029
+ More
ASJ26366.1 MF038031 ASJ26368.1 KQ459606 KPI91728.1 ODYU01003915 SOQ43266.1 KQ460779 KPJ12311.1 NWSH01003029 PCG67088.1 MF038036 ASJ26373.1 MF038026 ASJ26363.1 MF038033 ASJ26370.1 PCG67087.1 MF038027 ASJ26364.1 MF038025 ASJ26362.1 ODYU01006740 SOQ48866.1 MF038030 ASJ26367.1 MF038035 ASJ26372.1 MF038032 ASJ26369.1 KT358810 ALV66641.1 MF038024 ASJ26361.1 AK402113 BAM18735.1 AGBW02013216 OWR43859.1 MF038037 ASJ26374.1 MF038028 ASJ26365.1 LJIJ01000447 ODM97401.1 KT358808 ALV66639.1 AGBW02008548 OWR53055.1 MF038034 ASJ26371.1 CAEY01000030 KF981737 AHY81321.1 KF981736 AHY81319.1 LNIX01000020 OXA43944.1 NCKU01003151 NCKU01002640 NCKU01001078 RWS08070.1 RWS09121.1 RWS13159.1 LHCD01000027 KNY20625.1 FOQW01000018 SFJ43022.1 FOTK01000006 SFL51776.1 AKFK01000065 EIZ83823.1 LKKO01000019 KST60671.1 LJTP01000049 KPK65356.1 CP033231 AYO82577.1 FPCB01000005 SFU41101.1 BBUX01000052 GAN50939.1 CP001001 ACB25989.1 PEKQ01000092 PJI55431.1 FOZY01000002 SFS42367.1 CP029552 AWN54528.1 QEKZ01000031 PVY93979.1 MGQK01000094 OGP57509.1 NWMW01000002 PCD02189.1 LABX01000002 KMO41904.1 AAYA01000001 EBA10161.1 DNXT01000114 HBK45463.1 NJIH01000007 OWT59206.1 AUDA01000125 EPR15520.1 NETV01000003 PJG45351.1 FZOS01000023 SNS91314.1 ATDO01000050 EQB07481.1 PHHF01000073 PTD17464.1 FUYP01000024 SKB87024.1 CP005083 AMK21220.1 ATDP01000042 EQB18807.1 FUYM01000016 SKC09302.1 JFYY01000050 EZP66023.1 CP005191 AMK20679.1 JANF02000093 KER34691.1 AP017657 BAV66794.1 CP021913 AUC53845.1 CYSX01000013 CUJ78136.1 MJMN01000001 OMG93787.1 CP026124 AUT44835.1 FOZW01000010 SFT11459.1 QLNH01000445 RBL87084.1 CYTT01000021 CUJ79377.1 LN831029 CKI26839.1 PHIH01000025 PJM67232.1 CP017433 AOU96612.1 CP014060 AMG37948.1 CYUB01000030 CUK20320.1 CYTP01000003 CUJ12069.1 LWPK01000139 OAS88966.1 AAWL01000009 EAX47485.1 CP002780 AEG60882.1 AFRQ01000039 EGP46495.1 CP004393 AJE47628.1 CP019698 AQS58765.1 FRAR01000017 SHK58037.1 MGNS01000162 OGO33981.1 FQUY01000005 SHE72880.1 CP000612 ABO49895.1 MHBF01000088 OGV50990.1 CAOS01000003 CCO07430.1 AWQQ01000017 PHJ39559.1 RPQU01000440 RPI07673.1 KK037166 EWM10805.1 RQXT01000003 RRI06437.1 NSGG01000013 PBC07768.1 JRAS01000029 KGM44241.1 NEXF01000039 PSO08907.1 MGOC01000174 OGO51848.1 SADM01000010 RVV42596.1
ASJ26366.1 MF038031 ASJ26368.1 KQ459606 KPI91728.1 ODYU01003915 SOQ43266.1 KQ460779 KPJ12311.1 NWSH01003029 PCG67088.1 MF038036 ASJ26373.1 MF038026 ASJ26363.1 MF038033 ASJ26370.1 PCG67087.1 MF038027 ASJ26364.1 MF038025 ASJ26362.1 ODYU01006740 SOQ48866.1 MF038030 ASJ26367.1 MF038035 ASJ26372.1 MF038032 ASJ26369.1 KT358810 ALV66641.1 MF038024 ASJ26361.1 AK402113 BAM18735.1 AGBW02013216 OWR43859.1 MF038037 ASJ26374.1 MF038028 ASJ26365.1 LJIJ01000447 ODM97401.1 KT358808 ALV66639.1 AGBW02008548 OWR53055.1 MF038034 ASJ26371.1 CAEY01000030 KF981737 AHY81321.1 KF981736 AHY81319.1 LNIX01000020 OXA43944.1 NCKU01003151 NCKU01002640 NCKU01001078 RWS08070.1 RWS09121.1 RWS13159.1 LHCD01000027 KNY20625.1 FOQW01000018 SFJ43022.1 FOTK01000006 SFL51776.1 AKFK01000065 EIZ83823.1 LKKO01000019 KST60671.1 LJTP01000049 KPK65356.1 CP033231 AYO82577.1 FPCB01000005 SFU41101.1 BBUX01000052 GAN50939.1 CP001001 ACB25989.1 PEKQ01000092 PJI55431.1 FOZY01000002 SFS42367.1 CP029552 AWN54528.1 QEKZ01000031 PVY93979.1 MGQK01000094 OGP57509.1 NWMW01000002 PCD02189.1 LABX01000002 KMO41904.1 AAYA01000001 EBA10161.1 DNXT01000114 HBK45463.1 NJIH01000007 OWT59206.1 AUDA01000125 EPR15520.1 NETV01000003 PJG45351.1 FZOS01000023 SNS91314.1 ATDO01000050 EQB07481.1 PHHF01000073 PTD17464.1 FUYP01000024 SKB87024.1 CP005083 AMK21220.1 ATDP01000042 EQB18807.1 FUYM01000016 SKC09302.1 JFYY01000050 EZP66023.1 CP005191 AMK20679.1 JANF02000093 KER34691.1 AP017657 BAV66794.1 CP021913 AUC53845.1 CYSX01000013 CUJ78136.1 MJMN01000001 OMG93787.1 CP026124 AUT44835.1 FOZW01000010 SFT11459.1 QLNH01000445 RBL87084.1 CYTT01000021 CUJ79377.1 LN831029 CKI26839.1 PHIH01000025 PJM67232.1 CP017433 AOU96612.1 CP014060 AMG37948.1 CYUB01000030 CUK20320.1 CYTP01000003 CUJ12069.1 LWPK01000139 OAS88966.1 AAWL01000009 EAX47485.1 CP002780 AEG60882.1 AFRQ01000039 EGP46495.1 CP004393 AJE47628.1 CP019698 AQS58765.1 FRAR01000017 SHK58037.1 MGNS01000162 OGO33981.1 FQUY01000005 SHE72880.1 CP000612 ABO49895.1 MHBF01000088 OGV50990.1 CAOS01000003 CCO07430.1 AWQQ01000017 PHJ39559.1 RPQU01000440 RPI07673.1 KK037166 EWM10805.1 RQXT01000003 RRI06437.1 NSGG01000013 PBC07768.1 JRAS01000029 KGM44241.1 NEXF01000039 PSO08907.1 MGOC01000174 OGO51848.1 SADM01000010 RVV42596.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000094527 UP000015104 UP000198287 UP000285301 UP000036734 UP000199571 UP000199048 UP000004753 UP000053043 UP000278388 UP000199402 UP000032904 UP000006589 UP000198700 UP000246097 UP000246100 UP000218366 UP000035929 UP000005713 UP000214603 UP000019104 UP000229267 UP000198281 UP000015528 UP000241206 UP000190044 UP000060779 UP000015531 UP000189818 UP000023992 UP000060707 UP000028135 UP000218272 UP000232586 UP000041351 UP000187251 UP000236176 UP000199392 UP000043136 UP000059657 UP000229198 UP000192456 UP000060602 UP000039708 UP000040179 UP000078502 UP000005139 UP000009234 UP000004853 UP000031521 UP000189464 UP000183997 UP000179273 UP000184148 UP000001556 UP000176916 UP000009315 UP000030658 UP000273786 UP000217825 UP000242015 UP000288475
UP000094527 UP000015104 UP000198287 UP000285301 UP000036734 UP000199571 UP000199048 UP000004753 UP000053043 UP000278388 UP000199402 UP000032904 UP000006589 UP000198700 UP000246097 UP000246100 UP000218366 UP000035929 UP000005713 UP000214603 UP000019104 UP000229267 UP000198281 UP000015528 UP000241206 UP000190044 UP000060779 UP000015531 UP000189818 UP000023992 UP000060707 UP000028135 UP000218272 UP000232586 UP000041351 UP000187251 UP000236176 UP000199392 UP000043136 UP000059657 UP000229198 UP000192456 UP000060602 UP000039708 UP000040179 UP000078502 UP000005139 UP000009234 UP000004853 UP000031521 UP000189464 UP000183997 UP000179273 UP000184148 UP000001556 UP000176916 UP000009315 UP000030658 UP000273786 UP000217825 UP000242015 UP000288475
PRIDE
Pfam
PF00291 PALP
SUPFAM
SSF53686
SSF53686
ProteinModelPortal
H9JRG3
A0A0L7LB36
A0A0U3S5Y4
A0A220K8H3
A0A220K819
A0A194PFD1
+ More
A0A2H1VR62 A0A194R8W1 A0A2A4J5Y7 A0A220K8C9 A0A220K835 A0A220K8J3 A0A2A4J4L8 A0A220K8A8 A0A220K813 A0A2H1W7D9 A0A220K8E3 A0A220K8D1 A0A220K8F1 A0A0U3QZ45 A0A220K8B9 I4DLE5 A0A212EQW4 A0A220K8D8 A0A220K824 A0A1D2MWT8 A0A0U3SCD8 A0A212FH38 A0A220K8D2 T1KF23 A0A023ZRZ4 A0A226DGG8 A0A3S3NRW7 A0A0L6J4R0 A0A1I3RB50 A0A1I4IDD8 I9WTW2 A0A0V7Z7W8 A0A0S8G065 A0A3G2V492 A0A1I7FYA1 A0A0E9KEZ5 B1M078 A0A2M8VFB9 A0A1I6PQ87 A0A2U8X9Z7 A0A2U1E2Q1 A0A1F9AHE9 A0A2A4B330 A0A0J6T7G5 A3JXC5 A0A354Y7X9 A0A225MD82 W1KQD9 A0A2M8QIC4 A0A239IFX7 T0GU46 A0A2T4HNU1 A0A1T5ESV9 A0A126RG36 T0I0H1 A0A1T5GLM8 A0A031IXS0 A0A126RF03 A0A081SXB7 A0A1E1F8E0 A0A2K8WBZ3 A0A0M7MJC7 A0A1R1K2W7 A0A2I8DC77 A0A1I6VCR4 A0A365XMT5 A0A0M7MMQ9 A0A0D6IXP8 A0A2M9GRQ0 A0A1D8IJF5 A0A0X8P154 A0A0M7K288 A0A0M7HQQ6 A0A179T477 A1HQX6 F6DQI8 F7SZI9 A0A0B5DW83 A0A1S6IVG3 A0A1M6TM50 A0A1F8QFU6 A0A1M4VVD8 A4J492 A0A1G0YYH4 K8DXV2 A0A2C6ME27 A0A3N5KT09 W7S7N8 A0A3P3G6S4 A0A2A3DNU8 A0A0A0DZP8 A0A2R6CDD4 A0A1F8RW04 A0A3S2YAM0
A0A2H1VR62 A0A194R8W1 A0A2A4J5Y7 A0A220K8C9 A0A220K835 A0A220K8J3 A0A2A4J4L8 A0A220K8A8 A0A220K813 A0A2H1W7D9 A0A220K8E3 A0A220K8D1 A0A220K8F1 A0A0U3QZ45 A0A220K8B9 I4DLE5 A0A212EQW4 A0A220K8D8 A0A220K824 A0A1D2MWT8 A0A0U3SCD8 A0A212FH38 A0A220K8D2 T1KF23 A0A023ZRZ4 A0A226DGG8 A0A3S3NRW7 A0A0L6J4R0 A0A1I3RB50 A0A1I4IDD8 I9WTW2 A0A0V7Z7W8 A0A0S8G065 A0A3G2V492 A0A1I7FYA1 A0A0E9KEZ5 B1M078 A0A2M8VFB9 A0A1I6PQ87 A0A2U8X9Z7 A0A2U1E2Q1 A0A1F9AHE9 A0A2A4B330 A0A0J6T7G5 A3JXC5 A0A354Y7X9 A0A225MD82 W1KQD9 A0A2M8QIC4 A0A239IFX7 T0GU46 A0A2T4HNU1 A0A1T5ESV9 A0A126RG36 T0I0H1 A0A1T5GLM8 A0A031IXS0 A0A126RF03 A0A081SXB7 A0A1E1F8E0 A0A2K8WBZ3 A0A0M7MJC7 A0A1R1K2W7 A0A2I8DC77 A0A1I6VCR4 A0A365XMT5 A0A0M7MMQ9 A0A0D6IXP8 A0A2M9GRQ0 A0A1D8IJF5 A0A0X8P154 A0A0M7K288 A0A0M7HQQ6 A0A179T477 A1HQX6 F6DQI8 F7SZI9 A0A0B5DW83 A0A1S6IVG3 A0A1M6TM50 A0A1F8QFU6 A0A1M4VVD8 A4J492 A0A1G0YYH4 K8DXV2 A0A2C6ME27 A0A3N5KT09 W7S7N8 A0A3P3G6S4 A0A2A3DNU8 A0A0A0DZP8 A0A2R6CDD4 A0A1F8RW04 A0A3S2YAM0
PDB
5XOQ
E-value=2.60773e-39,
Score=406
Ontologies
KEGG
PATHWAY
00270
Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00920 Sulfur metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00920 Sulfur metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
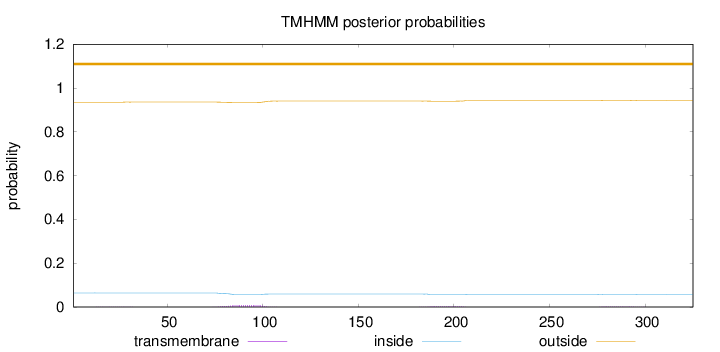
Topology
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2813
Exp number, first 60 AAs:
0.01095
Total prob of N-in:
0.06439
outside
1 - 325
Population Genetic Test Statistics
Pi
271.429166
Theta
200.120204
Tajima's D
1.121612
CLR
0.02281
CSRT
0.692565371731413
Interpretation
Uncertain
