Pre Gene Modal
BGIBMGA000829
Annotation
PREDICTED:_splicing_factor_3B_subunit_3_isoform_X1_[Amyelois_transitella]
Full name
Splicing factor 3B subunit 3
Alternative Name
Pre-mRNA-splicing factor SF3b 130 kDa subunit
Spliceosome-associated protein 130
Spliceosome-associated protein 130
Location in the cell
Cytoplasmic Reliability : 1.697 Nuclear Reliability : 1.217 PlasmaMembrane Reliability : 1.075
Sequence
CDS
ATGTATCTCTACAATTTAACACTGCAAGGTTCATCTGCCATAACACATGCCGTACATGGAAATTTCTCTGGAACTAAGCAACAGGAGATTATTATATCACGTGGCAAAACACTTGAATTGTTACGGCCAGATCCGAACACGGGAAAGGTCCACACATTAATGAAAATTGAAATATTTGGTGTAGTTCGGTCTATTATGGCTTTTCGTCTAACCGGAGGAACAAAAGATTACATTGTGGTTGGTTCTGATTCTGGACGTATCATCATTTTGGAGTACATCCCAACTAAGAACATTTTGGAGAAAGTCCATCAAGAAACATTTGGTAAATCTGGCTGTAGGAGAATAGTTCCTGGTCAATATCTATCAATTGATCCTAAAGGAAGAGCTGTTATGATTGGTGCAATTGAAAAACAGAAATTGGTATACATTTTGAATAGAGATGCAGAAGCTCGACTAACAATATCTTCACCACTTGAAGCTCACAAGTCAAACACACTAGTTTATCACATGGTTGGTGTTGATGTTGGATTTGAAAATCCCATGTTTGCATGTTTGGAAATTGATTATGAAGAAGCTGATTCAGATCCTACCGGCGAAGCTGCACAAAAAACTCAACAGACTTTAACATTTTACGAATTAGATCTTGGTTTGAATCATGTTGTTAGAAAGTACTCTGAACCTTTAGAAGAGCATGCAAACTTTCTCATAACTGTGCCTGGAGGAAATGATGGTCCTTCTGGTGTCCTTATATGCTCTGAAAATTATCTCACATACAAAAATTTGGGTGATCAACATGATATTAGGTGTCCTATACCTCGCAGAAGAAATGATTTAGATGATCCAGAAAGAGGAATGATCTTTGTTTGTTCTGCCACGCATAAAACTAAGTCCATGTTTTTCTTTTTGGCACAAACTGAACAGGGTGATATTTTCAAAATAACTATAGAAACTGATGAAGATATGGTCACTGAAATAAAGCTGAAATACTTTGACACTGTACCAGTGGCAACATCAATGTGTGTTCTTAAAACTGGATTTTTGTTTGTGGCTTGTGAATTTGGCAACCACTACTTATATCAGATAGCACATTTGGGTGATGAAGATGATGAACCAGAATTTAGCTCTGCTATGCCTCTAGAAGAGGGCGACACATTCTTTTTTGCACCAAGACCTTTGAGGAATTTAGTACTGGTAGATGAAATGGATTCTTTGTCACCAATATTAGCTTGCCATGTGGCAGATTTAGCTGGGGAAGATACTCCGCAAGTTTACTTGGCATGTGGGAGAGGCCCACGATCCTCCCTTAGAGCACTAAGACATGGTCTTGAAGTTGCTGAAATGGCTGTATCAGAGCTTCCAGGATCACCGAATGCTGTTTGGACAGTTCGACGAAATAAAGATGAGGAATACGATTCTTACATCATTGTGTCATTCGTTAATGCCACACTTGTTCTTTCCATCGGTGAGACTGTTGAAGAAGTCACTGACTCAGGATTTTTGGGAACTACACCTACACTTAGTTGTCATGCAATGGGTAATGACGCTCTAGTACAAGTATATCCTGATGGTATAAGACATATTCGTGCTGACAAACGTGTAAATGAATGGAAGGCTCCTGGAAAAAAATCTATAGTCAGATGTGCAGTCAATCAAAGACAAGTCGTCATTGCCCTAACTGGAGGGGAGTTGGTATATTTCGAAATGGACCCTACTGGACAGCTCAATGAATACACAGAAAGAAAGAAATTATCTTCAGATGTTTGTTGTATGGCATTAGGTTCAGTTGCAGCAGGGGAACAGAGAGCATGGTTTTTAGCAGTTGGTCTCAATGATAACACTGTCAGAATTATCTCTTTGGACCCAGCAGACTGTCTTTCACCACGTTCTATGCAAGCTCTTCCTGCTGGTGCAGAATCATTATGCATTATTGAGCAACCATTTGAATCTGGTGCAAAATCTGCCCTGCATTTAAACATTGGCTTAAGCAACGGGGTGCTGTTAAGAACTACCTTAGATTCTGTAAGCGGTGATCTTGCAGACACTAGAACTAGATACTTGGGGTCAAGACCTGTGAAGTTATTTAAAGTAAGGGTACAAGCTGCTGAAGCAGTACTAGCTGTGTCGTCTAGAACATGGCTAGGATATCACTATCAAAATCGTTTTCACCTTACACCTCTATCTTATGAATGTTTGGAATATGCAGCAGGATTTAGTTCAGAACAATGTACTGAAGGCATTGTAGCCATATCTTCCAACACTCTGAGAATTTTGGCTTTAGAAAAATTGGGAGCTGTATTCAATCAAACATTTGTCCCTTTAGAATACACACCAAGAAAGTTTATAATAAATTCTGACAACAATCACATTATAGTTTTAGAAACTGATCACAATGCCTACACAGAGGAAATGAAAAAACACAGAAGAATTCAAATGGCCCAAGAAATGCGTGAAGCTGCTGCTGGAGGTGCTCCTGAAGAACAGCAACTAGCAAATGAAATGGCAGATGCTTTTCTATCTGATACTTTGCCTGAATACATTTTTTCTTCACCTAAAGCTGGAGCAGGCATGTGGGCATCTCTAATTCGTGTTGTTGACATGGGTATTGGTGGGGGTCAACCCAATACATTATTTAGACTTCCACTGGAGCAAAACGAAGCAGCTGTTTCTTTGTGCATAGTAAGATGGGCTGCACATGCTGAACATGCACAACCACATCTTGTGGTTGGTGTTGCAAAGGATTTGATACTGTCTCCTCGTTCTTGCACTGAAGGCAGTCTACATGTTTATAAGATATATGGCAACACTGGAAAATTAGAACTTGTTCACAAAACTCCAGTAGATGAATATCCTGGTGCTATAGCAGCATTTAATGGTAGACTACTAGCTGGTGTTGGACGCATGTTAAGATTATATGATATTGGACGACGAAAACTCCTACGTAAATGTGAAAATAGACATATTCCAAATTTAATAGCAGATATAAAAACGATTGGACAGAGAATTTTTGTTTCTGATGTCCAAGAATCTGTATTTTGTGTCAAGCATAAGAAAAGAGAGAATCAATTAATTATTTTTGCTGATGATACCAATCCTAGATGGATAACAAATAGCTGTATTCTTGATTATGACACTATAGCAGTATCTGACAAATTTGGTAATGTGGCTATAATGCGGTTACCACAATCTGTAAGTGATGATGTTGATGAAGATCCTACTGGAAACAAAGCACTTTGGGACAGAGGTTTACTTAATGGTGCTTCACAAAAGGGAGATGTTGTGGTCAATTTTCATGTTGGTGAAACTGTAACCTCTCTACAGAGAGCAACATTAATTCCTGGTGGTTCTGAAGCACTACTATATGCAACAATTAGTGGGTCTCTTGGCGTTCTACTACCGTTTACATCAAGAGAAGATCATGACTTCTTTCAACATCTAGAGATGCATATGAGAAGTGAAAATTCACCATTATGTGGGCGTGACCATTTATCTTTTAGAAGTTATTACTACCCAGTAAAGAATGTGATTGACGGAGATCTATGTGAACAGTTTAACTCCCTAGACCCTGGAAAACAAAAAGCCATCGCAGGTGATTTGGAACGTACTCCCGCTGAAGTATCAAAAAAACTTGAAGACATTCGAACTAGATATGCGTTCTAA
Protein
MYLYNLTLQGSSAITHAVHGNFSGTKQQEIIISRGKTLELLRPDPNTGKVHTLMKIEIFGVVRSIMAFRLTGGTKDYIVVGSDSGRIIILEYIPTKNILEKVHQETFGKSGCRRIVPGQYLSIDPKGRAVMIGAIEKQKLVYILNRDAEARLTISSPLEAHKSNTLVYHMVGVDVGFENPMFACLEIDYEEADSDPTGEAAQKTQQTLTFYELDLGLNHVVRKYSEPLEEHANFLITVPGGNDGPSGVLICSENYLTYKNLGDQHDIRCPIPRRRNDLDDPERGMIFVCSATHKTKSMFFFLAQTEQGDIFKITIETDEDMVTEIKLKYFDTVPVATSMCVLKTGFLFVACEFGNHYLYQIAHLGDEDDEPEFSSAMPLEEGDTFFFAPRPLRNLVLVDEMDSLSPILACHVADLAGEDTPQVYLACGRGPRSSLRALRHGLEVAEMAVSELPGSPNAVWTVRRNKDEEYDSYIIVSFVNATLVLSIGETVEEVTDSGFLGTTPTLSCHAMGNDALVQVYPDGIRHIRADKRVNEWKAPGKKSIVRCAVNQRQVVIALTGGELVYFEMDPTGQLNEYTERKKLSSDVCCMALGSVAAGEQRAWFLAVGLNDNTVRIISLDPADCLSPRSMQALPAGAESLCIIEQPFESGAKSALHLNIGLSNGVLLRTTLDSVSGDLADTRTRYLGSRPVKLFKVRVQAAEAVLAVSSRTWLGYHYQNRFHLTPLSYECLEYAAGFSSEQCTEGIVAISSNTLRILALEKLGAVFNQTFVPLEYTPRKFIINSDNNHIIVLETDHNAYTEEMKKHRRIQMAQEMREAAAGGAPEEQQLANEMADAFLSDTLPEYIFSSPKAGAGMWASLIRVVDMGIGGGQPNTLFRLPLEQNEAAVSLCIVRWAAHAEHAQPHLVVGVAKDLILSPRSCTEGSLHVYKIYGNTGKLELVHKTPVDEYPGAIAAFNGRLLAGVGRMLRLYDIGRRKLLRKCENRHIPNLIADIKTIGQRIFVSDVQESVFCVKHKKRENQLIIFADDTNPRWITNSCILDYDTIAVSDKFGNVAIMRLPQSVSDDVDEDPTGNKALWDRGLLNGASQKGDVVVNFHVGETVTSLQRATLIPGGSEALLYATISGSLGVLLPFTSREDHDFFQHLEMHMRSENSPLCGRDHLSFRSYYYPVKNVIDGDLCEQFNSLDPGKQKAIAGDLERTPAEVSKKLEDIRTRYAF
Summary
Description
Involved in pre-mRNA splicing as a component of the splicing factor SF3B complex, a constituent of the spliceosome. SF3B complex is required for 'A' complex assembly formed by the stable binding of U2 snRNP to the branchpoint sequence (BPS) in pre-mRNA. Sequence independent binding of SF3A/SF3B complex upstream of the branch site is essential, it may anchor U2 snRNP to the pre-mRNA. May also be involved in the assembly of the 'E' complex. Belongs also to the minor U12-dependent spliceosome, which is involved in the splicing of rare class of nuclear pre-mRNA intron.
Subunit
Identified in the spliceosome A complex; remains associated with the spliceosome throughout the splicing process. Component of the spliceosome B complex. Identified in the spliceosome C complex. Identified in the spliceosome E complex. Component of the U11/U12 snRNPs that are part of the U12-type spliceosome. Component of splicing factor SF3B complex which is composed of at least eight subunits; SF3B1, SF3B2, SF3B3, SF3B4, SF3B5, SF3B6, PHF5A and DDX42. SF3B associates with the splicing factor SF3A and a 12S RNA unit to form the U2 small nuclear ribonucleoproteins complex (U2 snRNP).
Identified in the spliceosome A complex; remains associated with the spliceosome throughout the splicing process. Component of the spliceosome B complex. Identified in the spliceosome C complex. Identified in the spliceosome E complex. Component of the U11/U12 snRNPs that are part of the U12-type spliceosome. Component of splicing factor SF3B complex which is composed of at least eight subunits; SF3B1, SF3B2, SF3B3, SF3B4, SF3B5, SF3B6, PHF5A and DDX42. SF3B associates with the splicing factor SF3A and a 12S RNA unit to form the U2 small nuclear ribonucleoproteins complex (U2 snRNP). Interaction between SF3B3 and SF3B1 is tighter than the interaction between SF3B3 and SF3B2. Within the SF3B complex interacts directly with SF3B1 (via HEAT domain), SF3B5 and PHF5A. The SF3B complex composed of SF3B1, SF3B2, SF3B3, SF3B4, SF3B5, SF3B6 and PHF5A interacts with U2AF2. Associates with the STAGA transcription coactivator-HAT complex. Interacts with SUPT3H. Interacts with TAF3.
Identified in the spliceosome A complex; remains associated with the spliceosome throughout the splicing process. Component of the spliceosome B complex. Identified in the spliceosome C complex. Identified in the spliceosome E complex. Component of the U11/U12 snRNPs that are part of the U12-type spliceosome. Component of splicing factor SF3B complex which is composed of at least eight subunits; SF3B1, SF3B2, SF3B3, SF3B4, SF3B5, SF3B6, PHF5A and DDX42. SF3B associates with the splicing factor SF3A and a 12S RNA unit to form the U2 small nuclear ribonucleoproteins complex (U2 snRNP). Interaction between SF3B3 and SF3B1 is tighter than the interaction between SF3B3 and SF3B2. Within the SF3B complex interacts directly with SF3B1 (via HEAT domain), SF3B5 and PHF5A. The SF3B complex composed of SF3B1, SF3B2, SF3B3, SF3B4, SF3B5, SF3B6 and PHF5A interacts with U2AF2. Associates with the STAGA transcription coactivator-HAT complex. Interacts with SUPT3H. Interacts with TAF3.
Similarity
Belongs to the RSE1 family.
Keywords
Complete proteome
mRNA processing
mRNA splicing
Nucleus
Reference proteome
Spliceosome
Alternative splicing
Phosphoprotein
Feature
chain Splicing factor 3B subunit 3
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9IU99
A0A194R3C6
A0A2H1W0W3
A0A2A4JMG5
A0A212ELF1
A0A0L7LDM8
+ More
A0A1Y1LLA2 E2BJM4 A0A026W5W7 A0A0N0BEW6 A0A067RIM3 E0VUC3 A0A2J7PDD4 A0A0L7QUK3 A0A151IBP3 A0A232F8J7 A0A154PRA3 E9IIH8 A0A158NM51 F4WHN1 A0A151JWJ7 A0A151WIT1 A0A182H7T4 D6WGP5 Q17HE5 A0A1L8DX60 A0A195B599 B0WIA3 A0A1Q3F4Y3 A0A1B6EDX5 A0A084WKK5 A0A182JGL5 A0A0J7KJK7 A0A182N9M1 A0A151J7A9 A0A182TWQ5 A0A182K875 A0A182Y121 A0A182HLY5 A0A182V1X0 A0A182X538 A0A182LJE3 Q7Q722 W5J9Q8 A0A182PWA4 A0A182FG96 A0A182QWR2 A0A182MD81 A0A182WKD8 A0A182RMG1 A0A1W4Y195 W5U7N2 A0A3B4CD56 W5K7B9 A0A3Q3SIF7 W5MLC5 A0A0B8RTN8 H9G5S6 A0A0F7Z5C9 A0A3B3RDG2 A0A3Q1IPS3 A0A1L8GLL3 A0A1W7RDD3 T1E4U0 I3JYZ8 A0A0P7XEV7 A0A3B4XZR5 A0A1S3KM32 A0A3B4VFX3 C0H8V4 V9K9Y4 A0A087YCG5 A0A3Q1AQY3 A0A3B3UE66 A0A3P8U5I8 A0A3Q2QUK2 A0A3B4F3V3 F6ZI40 A0A3Q2E046 A0A3B3XQ26 A0A3P9J277 A0A3B3IK37 A0A3P9L6Q4 A0A3P9BWZ9 Q1LVE8 A0A0F7Z4R7 A0A091GKD3 G1MS46 A0A3B3C4Z4 A0A1D6UPS9 G3HAF4 E9PT66 B2RSV4 L5MD50 S7MMM5 A0A224X5K2 Q921M3 A0A2J8VW45 A0A2I4C7V8 A0A3Q3QF45
A0A1Y1LLA2 E2BJM4 A0A026W5W7 A0A0N0BEW6 A0A067RIM3 E0VUC3 A0A2J7PDD4 A0A0L7QUK3 A0A151IBP3 A0A232F8J7 A0A154PRA3 E9IIH8 A0A158NM51 F4WHN1 A0A151JWJ7 A0A151WIT1 A0A182H7T4 D6WGP5 Q17HE5 A0A1L8DX60 A0A195B599 B0WIA3 A0A1Q3F4Y3 A0A1B6EDX5 A0A084WKK5 A0A182JGL5 A0A0J7KJK7 A0A182N9M1 A0A151J7A9 A0A182TWQ5 A0A182K875 A0A182Y121 A0A182HLY5 A0A182V1X0 A0A182X538 A0A182LJE3 Q7Q722 W5J9Q8 A0A182PWA4 A0A182FG96 A0A182QWR2 A0A182MD81 A0A182WKD8 A0A182RMG1 A0A1W4Y195 W5U7N2 A0A3B4CD56 W5K7B9 A0A3Q3SIF7 W5MLC5 A0A0B8RTN8 H9G5S6 A0A0F7Z5C9 A0A3B3RDG2 A0A3Q1IPS3 A0A1L8GLL3 A0A1W7RDD3 T1E4U0 I3JYZ8 A0A0P7XEV7 A0A3B4XZR5 A0A1S3KM32 A0A3B4VFX3 C0H8V4 V9K9Y4 A0A087YCG5 A0A3Q1AQY3 A0A3B3UE66 A0A3P8U5I8 A0A3Q2QUK2 A0A3B4F3V3 F6ZI40 A0A3Q2E046 A0A3B3XQ26 A0A3P9J277 A0A3B3IK37 A0A3P9L6Q4 A0A3P9BWZ9 Q1LVE8 A0A0F7Z4R7 A0A091GKD3 G1MS46 A0A3B3C4Z4 A0A1D6UPS9 G3HAF4 E9PT66 B2RSV4 L5MD50 S7MMM5 A0A224X5K2 Q921M3 A0A2J8VW45 A0A2I4C7V8 A0A3Q3QF45
Pubmed
19121390
26354079
22118469
26227816
28004739
20798317
+ More
24508170 24845553 20566863 28648823 21282665 21347285 21719571 26483478 18362917 19820115 17510324 24438588 25244985 20966253 12364791 14747013 17210077 20920257 23761445 23127152 25329095 25476704 21881562 29240929 27762356 26358130 23758969 25186727 20433749 24402279 20431018 17554307 23594743 20838655 29451363 15592404 21804562 15057822 12040188 15489334 16141072 14621295 21183079
24508170 24845553 20566863 28648823 21282665 21347285 21719571 26483478 18362917 19820115 17510324 24438588 25244985 20966253 12364791 14747013 17210077 20920257 23761445 23127152 25329095 25476704 21881562 29240929 27762356 26358130 23758969 25186727 20433749 24402279 20431018 17554307 23594743 20838655 29451363 15592404 21804562 15057822 12040188 15489334 16141072 14621295 21183079
EMBL
BABH01042095
KQ460779
KPJ12308.1
ODYU01005620
SOQ46653.1
NWSH01001104
+ More
PCG72590.1 AGBW02014084 OWR42297.1 JTDY01001566 KOB73505.1 GEZM01052568 JAV74432.1 GL448604 EFN84082.1 KK107390 EZA51467.1 KQ435821 KOX72481.1 KK852609 KDR20301.1 DS235785 EEB16979.1 NEVH01026391 PNF14333.1 KQ414735 KOC62300.1 KQ978079 KYM97150.1 NNAY01000655 OXU27161.1 KQ435078 KZC14455.1 GL763491 EFZ19622.1 ADTU01020258 GL888167 EGI66238.1 KQ981663 KYN38503.1 KQ983075 KYQ47757.1 JXUM01116879 JXUM01116880 KQ566028 KXJ70446.1 KQ971329 EFA01110.1 CH477250 EAT46061.1 GFDF01003085 JAV10999.1 KQ976604 KYM79364.1 DS231945 EDS28329.1 GFDL01012472 JAV22573.1 GEDC01001175 JAS36123.1 ATLV01024123 KE525349 KFB50749.1 LBMM01006658 KMQ90416.1 KQ979737 KYN19373.1 APCN01004272 AAAB01008960 EAA11859.1 ADMH02002078 ETN59484.1 AXCN02000984 AXCM01002538 JT406416 AHH37584.1 AHAT01020841 GBSH01002520 JAG66506.1 AAWZ02029915 AAWZ02029916 GBEX01003162 JAI11398.1 CM004472 OCT84666.1 GDAY02002283 JAV49149.1 GAAZ01002331 JAA95612.1 AERX01003723 AERX01003724 JARO02001941 KPP74015.1 BT058760 ACN10473.1 JW861997 AFO94514.1 AYCK01011957 AAMC01003452 AAMC01003453 AAMC01003454 AAMC01003455 AAMC01003456 AAMC01003457 AAMC01003458 BX784024 BC047171 GBEW01000961 JAI09404.1 KL448320 KFO82820.1 AADN05000561 JH000251 EGW04118.1 AC112801 BC139015 BC139016 CH466525 AAI39016.1 EDL11472.1 KB101785 ELK36185.1 KE161800 EPQ05519.1 GFTR01008670 JAW07756.1 AK085705 AK088268 AK129035 AK147914 BC011412 BC031197 BC042580 NDHI03003408 PNJ61752.1
PCG72590.1 AGBW02014084 OWR42297.1 JTDY01001566 KOB73505.1 GEZM01052568 JAV74432.1 GL448604 EFN84082.1 KK107390 EZA51467.1 KQ435821 KOX72481.1 KK852609 KDR20301.1 DS235785 EEB16979.1 NEVH01026391 PNF14333.1 KQ414735 KOC62300.1 KQ978079 KYM97150.1 NNAY01000655 OXU27161.1 KQ435078 KZC14455.1 GL763491 EFZ19622.1 ADTU01020258 GL888167 EGI66238.1 KQ981663 KYN38503.1 KQ983075 KYQ47757.1 JXUM01116879 JXUM01116880 KQ566028 KXJ70446.1 KQ971329 EFA01110.1 CH477250 EAT46061.1 GFDF01003085 JAV10999.1 KQ976604 KYM79364.1 DS231945 EDS28329.1 GFDL01012472 JAV22573.1 GEDC01001175 JAS36123.1 ATLV01024123 KE525349 KFB50749.1 LBMM01006658 KMQ90416.1 KQ979737 KYN19373.1 APCN01004272 AAAB01008960 EAA11859.1 ADMH02002078 ETN59484.1 AXCN02000984 AXCM01002538 JT406416 AHH37584.1 AHAT01020841 GBSH01002520 JAG66506.1 AAWZ02029915 AAWZ02029916 GBEX01003162 JAI11398.1 CM004472 OCT84666.1 GDAY02002283 JAV49149.1 GAAZ01002331 JAA95612.1 AERX01003723 AERX01003724 JARO02001941 KPP74015.1 BT058760 ACN10473.1 JW861997 AFO94514.1 AYCK01011957 AAMC01003452 AAMC01003453 AAMC01003454 AAMC01003455 AAMC01003456 AAMC01003457 AAMC01003458 BX784024 BC047171 GBEW01000961 JAI09404.1 KL448320 KFO82820.1 AADN05000561 JH000251 EGW04118.1 AC112801 BC139015 BC139016 CH466525 AAI39016.1 EDL11472.1 KB101785 ELK36185.1 KE161800 EPQ05519.1 GFTR01008670 JAW07756.1 AK085705 AK088268 AK129035 AK147914 BC011412 BC031197 BC042580 NDHI03003408 PNJ61752.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000037510
UP000008237
+ More
UP000053097 UP000053105 UP000027135 UP000009046 UP000235965 UP000053825 UP000078542 UP000215335 UP000076502 UP000005205 UP000007755 UP000078541 UP000075809 UP000069940 UP000249989 UP000007266 UP000008820 UP000078540 UP000002320 UP000030765 UP000075880 UP000036403 UP000075884 UP000078492 UP000075902 UP000075881 UP000076408 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000000673 UP000075885 UP000069272 UP000075886 UP000075883 UP000075920 UP000075900 UP000192224 UP000221080 UP000261440 UP000018467 UP000261640 UP000018468 UP000001646 UP000261540 UP000265040 UP000186698 UP000005207 UP000034805 UP000261360 UP000087266 UP000261420 UP000028760 UP000257160 UP000261500 UP000265080 UP000265000 UP000261460 UP000008143 UP000265020 UP000261480 UP000265200 UP000001038 UP000265180 UP000265160 UP000000437 UP000053760 UP000001645 UP000261560 UP000000539 UP000001075 UP000002494 UP000000589 UP000192220 UP000261600
UP000053097 UP000053105 UP000027135 UP000009046 UP000235965 UP000053825 UP000078542 UP000215335 UP000076502 UP000005205 UP000007755 UP000078541 UP000075809 UP000069940 UP000249989 UP000007266 UP000008820 UP000078540 UP000002320 UP000030765 UP000075880 UP000036403 UP000075884 UP000078492 UP000075902 UP000075881 UP000076408 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000000673 UP000075885 UP000069272 UP000075886 UP000075883 UP000075920 UP000075900 UP000192224 UP000221080 UP000261440 UP000018467 UP000261640 UP000018468 UP000001646 UP000261540 UP000265040 UP000186698 UP000005207 UP000034805 UP000261360 UP000087266 UP000261420 UP000028760 UP000257160 UP000261500 UP000265080 UP000265000 UP000261460 UP000008143 UP000265020 UP000261480 UP000265200 UP000001038 UP000265180 UP000265160 UP000000437 UP000053760 UP000001645 UP000261560 UP000000539 UP000001075 UP000002494 UP000000589 UP000192220 UP000261600
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9IU99
A0A194R3C6
A0A2H1W0W3
A0A2A4JMG5
A0A212ELF1
A0A0L7LDM8
+ More
A0A1Y1LLA2 E2BJM4 A0A026W5W7 A0A0N0BEW6 A0A067RIM3 E0VUC3 A0A2J7PDD4 A0A0L7QUK3 A0A151IBP3 A0A232F8J7 A0A154PRA3 E9IIH8 A0A158NM51 F4WHN1 A0A151JWJ7 A0A151WIT1 A0A182H7T4 D6WGP5 Q17HE5 A0A1L8DX60 A0A195B599 B0WIA3 A0A1Q3F4Y3 A0A1B6EDX5 A0A084WKK5 A0A182JGL5 A0A0J7KJK7 A0A182N9M1 A0A151J7A9 A0A182TWQ5 A0A182K875 A0A182Y121 A0A182HLY5 A0A182V1X0 A0A182X538 A0A182LJE3 Q7Q722 W5J9Q8 A0A182PWA4 A0A182FG96 A0A182QWR2 A0A182MD81 A0A182WKD8 A0A182RMG1 A0A1W4Y195 W5U7N2 A0A3B4CD56 W5K7B9 A0A3Q3SIF7 W5MLC5 A0A0B8RTN8 H9G5S6 A0A0F7Z5C9 A0A3B3RDG2 A0A3Q1IPS3 A0A1L8GLL3 A0A1W7RDD3 T1E4U0 I3JYZ8 A0A0P7XEV7 A0A3B4XZR5 A0A1S3KM32 A0A3B4VFX3 C0H8V4 V9K9Y4 A0A087YCG5 A0A3Q1AQY3 A0A3B3UE66 A0A3P8U5I8 A0A3Q2QUK2 A0A3B4F3V3 F6ZI40 A0A3Q2E046 A0A3B3XQ26 A0A3P9J277 A0A3B3IK37 A0A3P9L6Q4 A0A3P9BWZ9 Q1LVE8 A0A0F7Z4R7 A0A091GKD3 G1MS46 A0A3B3C4Z4 A0A1D6UPS9 G3HAF4 E9PT66 B2RSV4 L5MD50 S7MMM5 A0A224X5K2 Q921M3 A0A2J8VW45 A0A2I4C7V8 A0A3Q3QF45
A0A1Y1LLA2 E2BJM4 A0A026W5W7 A0A0N0BEW6 A0A067RIM3 E0VUC3 A0A2J7PDD4 A0A0L7QUK3 A0A151IBP3 A0A232F8J7 A0A154PRA3 E9IIH8 A0A158NM51 F4WHN1 A0A151JWJ7 A0A151WIT1 A0A182H7T4 D6WGP5 Q17HE5 A0A1L8DX60 A0A195B599 B0WIA3 A0A1Q3F4Y3 A0A1B6EDX5 A0A084WKK5 A0A182JGL5 A0A0J7KJK7 A0A182N9M1 A0A151J7A9 A0A182TWQ5 A0A182K875 A0A182Y121 A0A182HLY5 A0A182V1X0 A0A182X538 A0A182LJE3 Q7Q722 W5J9Q8 A0A182PWA4 A0A182FG96 A0A182QWR2 A0A182MD81 A0A182WKD8 A0A182RMG1 A0A1W4Y195 W5U7N2 A0A3B4CD56 W5K7B9 A0A3Q3SIF7 W5MLC5 A0A0B8RTN8 H9G5S6 A0A0F7Z5C9 A0A3B3RDG2 A0A3Q1IPS3 A0A1L8GLL3 A0A1W7RDD3 T1E4U0 I3JYZ8 A0A0P7XEV7 A0A3B4XZR5 A0A1S3KM32 A0A3B4VFX3 C0H8V4 V9K9Y4 A0A087YCG5 A0A3Q1AQY3 A0A3B3UE66 A0A3P8U5I8 A0A3Q2QUK2 A0A3B4F3V3 F6ZI40 A0A3Q2E046 A0A3B3XQ26 A0A3P9J277 A0A3B3IK37 A0A3P9L6Q4 A0A3P9BWZ9 Q1LVE8 A0A0F7Z4R7 A0A091GKD3 G1MS46 A0A3B3C4Z4 A0A1D6UPS9 G3HAF4 E9PT66 B2RSV4 L5MD50 S7MMM5 A0A224X5K2 Q921M3 A0A2J8VW45 A0A2I4C7V8 A0A3Q3QF45
PDB
6QX9
E-value=0,
Score=5174
Ontologies
GO
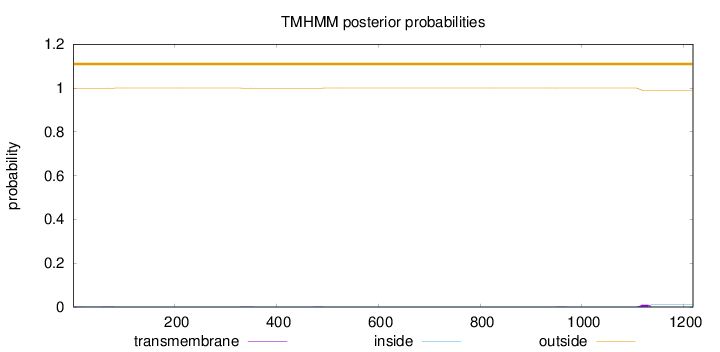
Topology
Subcellular location
Nucleus
Length:
1218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24805
Exp number, first 60 AAs:
0.00407
Total prob of N-in:
0.00077
outside
1 - 1218
Population Genetic Test Statistics
Pi
206.043197
Theta
232.490685
Tajima's D
-1.492934
CLR
1220.820913
CSRT
0.063896805159742
Interpretation
Uncertain
