Gene
KWMTBOMO06763 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012030
Annotation
PREDICTED:_serrate_RNA_effector_molecule_homolog_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.471
Sequence
CDS
ATGGAGATCCTGAAACAAGTGGCTAACGCACAGGTCGTGTTCACTGCTGCGGTGGTATCCGTTGTTTTAGTTTGTGCGGCGTTGGTATTTGTTTTTGGGTTCCGTTCTGCTGAACCGCCGCAATTCGACAAGTTGCCGCTAGTATTGGACGACAAGAAATCTGCCAAAGCCAAGAATAAAAAAAAGGAGAAGAAGTCTTCACCCAACCGATCTACATCTGATGATGTGAAGACCAAGGCTGATTCCTCCAAGAAATCTCCATCTAAGGAGAAAAAGGAGGAAAAAGTAAAGGAAGCTGACAAGCCTAAGCCTAAGGAAAAATCTGTTGAACCCAAGTTAGCTAAAAAAGAAGCAGTTATTGAAAAGAAAAATAGAAAGAATAAGGCTGGTGCAGAAGTTGAAAAACCTGCTGACTTTGATGATGGTAATTGGGAAGAGGTACCTAAAAAGAATGATAAGAAAGGAGAAAAAAAGAAAGTAAAGGCTCCAGAGGAAAAACAGAAGGAAAAAGAAAAGAAAAAAGAAAGTCCCATCAAGAAGGTTAAAAAAGCAAAGGATACAGATGTTGAAGCTGCTAGACCTAATGAAGATTCAAGTGAGAAAGTTAAAATTGTTAGTGTGAAAGGCCCTAACTTAGATGCGAATGTAACAAAAGCACTTCAAGAACAAGTGAAAGAATTTGAACGTGCAATGAAAGAGGCTCAACGCCGAGACCAGGGAATTGTGAATGATGAAGGATCCAATGAAATTGAGGAGCTGACTGACGTTAAAGATCTCAGAAGCAATAAAAAAAAGGAAAATAAAGAGAAACAAATAAAGAAAAAGGCTGGTGAGATCAAAGCAGATTCTGTAGAGAAGGAAAACTTGGAGCCTGAGAAACAGGAGGAAAATCAGAATGCACCAGCTTTTGATGAGCTTGGTGATACCTGGACAGAATCAAAAACATCCAAAAAAGGAAAAAAGAAAGCTCGCAAAGACCAGTGA
Protein
MEILKQVANAQVVFTAAVVSVVLVCAALVFVFGFRSAEPPQFDKLPLVLDDKKSAKAKNKKKEKKSSPNRSTSDDVKTKADSSKKSPSKEKKEEKVKEADKPKPKEKSVEPKLAKKEAVIEKKNRKNKAGAEVEKPADFDDGNWEEVPKKNDKKGEKKKVKAPEEKQKEKEKKKESPIKKVKKAKDTDVEAARPNEDSSEKVKIVSVKGPNLDANVTKALQEQVKEFERAMKEAQRRDQGIVNDEGSNEIEELTDVKDLRSNKKKENKEKQIKKKAGEIKADSVEKENLEPEKQEENQNAPAFDELGDTWTESKTSKKGKKKARKDQ
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00085 Thioredoxin
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
Ontologies
KEGG
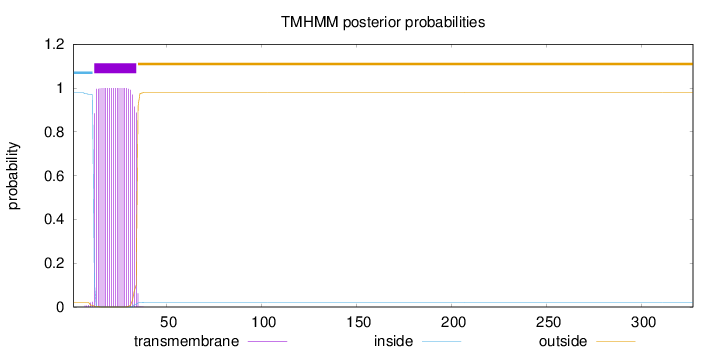
Topology
Length:
327
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.77245
Exp number, first 60 AAs:
22.77245
Total prob of N-in:
0.97981
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 327
Population Genetic Test Statistics
Pi
151.347676
Theta
137.280368
Tajima's D
0.153205
CLR
0
CSRT
0.412779361031948
Interpretation
Uncertain
