Pre Gene Modal
BGIBMGA012029
Annotation
mitochondrial_thioredoxin_2_[Bombyx_mori]
Full name
Thioredoxin
Location in the cell
Mitochondrial Reliability : 1.153
Sequence
CDS
ATGCTTACCAAAAATATTACTAACTTATTCATTCGAAATTGTACATTAAAAAAAAATTATGGGTTTTTGAGAAATTTCTCTCTAACTGCGTCGAAAAACGACATTGTGAAAATCCAGAGTACTGATGACTTCAAAGAAAAAGTAATTAATAGTAAAGTTCCCGTCGTTGTCGATTTCTTTGCAACGTGGTGCAACCCATGTAGACTACTTACTCCTCGCCTTGAGTCTATAATAGCAGAAAGTAAAGGCAAGGTGGTTTTGGCAAAAGTAGACATTGATGAACAAACTGATTTAGCATTAGATTATGAAGTAAGCTCTGTACCAGTCTTAGTAGCGATAAAGAATGGGAAAGTACAAAACCGTTTAGTTGGACTGCAAGATACTGATAAACTACGCAAGTGGATTGAACAGTTTGCTTCTGAAGAAACTAAAGCAGAAATAAAAGCTTAA
Protein
MLTKNITNLFIRNCTLKKNYGFLRNFSLTASKNDIVKIQSTDDFKEKVINSKVPVVVDFFATWCNPCRLLTPRLESIIAESKGKVVLAKVDIDEQTDLALDYEVSSVPVLVAIKNGKVQNRLVGLQDTDKLRKWIEQFASEETKAEIKA
Summary
Similarity
Belongs to the thioredoxin family.
Uniprot
Q2F5J9
I4DKF5
A0A194R3C0
A0A0L7LDN2
A0A3S2M9J8
A0A2A4JKL4
+ More
A0A0A0VA40 A0A212ELD1 A0A2H1WZ26 A0A2P8ZKG6 A0A224XNX0 A0A2J7RGR7 A0A023FAV9 A0A161N1J1 B0WA02 T1DH89 T1HM49 A0A0P4VHF6 A0A0A9ZB97 A0A182MSJ7 A0A182YMK2 A0A1Q3FJ16 A0A182WJS9 A0A084VC58 A0A2M4AA76 A0A2M4AMK8 A0A1A9Z673 A0A182UA47 A0A182PDN6 A0A182VF20 A0A182XLI1 Q7QJ17 A0A182I2P7 A0A182NJY5 A0A182K161 A0A2M4C2Y0 A0A182FGK8 A0A2M4C2X2 T1DNH7 A0A1B0AU90 A0A2M3ZI13 Q5MIS6 A0A182G788 A0A023EGR0 R4WPK4 A0A182QER1 Q16W35 A0A2M3Z9Q5 A0A2M3Z830 A0A182R3K2 A0A182J5U0 W5JKW7 A0A1J1ISX0 A0A2R7W9U2 A0A1A9VTX9 E0VVV8 E2BYJ1 A0A1I8NEF7 A0A026X324 B4N364 A0A1I8PYY6 A0A0L0BQY9 A0A1B0DBV4 A0A1W4W3Q4 B3M6P1 A0A1B0G8M7 A0A1B0DGC8 A0A336KEN9 A0A1L8EEM8 B4LBV9 A0A0K8TS39 B4J0H9 B4H5V2 A0A1L8EEY1 Q2LYS4 A0A1B6CNU0 A0A1B6H879 A0A1B6MJC6 A0A1S3CX50 A0A3B0KGR9 W8C243 A0A1L8E035
A0A0A0VA40 A0A212ELD1 A0A2H1WZ26 A0A2P8ZKG6 A0A224XNX0 A0A2J7RGR7 A0A023FAV9 A0A161N1J1 B0WA02 T1DH89 T1HM49 A0A0P4VHF6 A0A0A9ZB97 A0A182MSJ7 A0A182YMK2 A0A1Q3FJ16 A0A182WJS9 A0A084VC58 A0A2M4AA76 A0A2M4AMK8 A0A1A9Z673 A0A182UA47 A0A182PDN6 A0A182VF20 A0A182XLI1 Q7QJ17 A0A182I2P7 A0A182NJY5 A0A182K161 A0A2M4C2Y0 A0A182FGK8 A0A2M4C2X2 T1DNH7 A0A1B0AU90 A0A2M3ZI13 Q5MIS6 A0A182G788 A0A023EGR0 R4WPK4 A0A182QER1 Q16W35 A0A2M3Z9Q5 A0A2M3Z830 A0A182R3K2 A0A182J5U0 W5JKW7 A0A1J1ISX0 A0A2R7W9U2 A0A1A9VTX9 E0VVV8 E2BYJ1 A0A1I8NEF7 A0A026X324 B4N364 A0A1I8PYY6 A0A0L0BQY9 A0A1B0DBV4 A0A1W4W3Q4 B3M6P1 A0A1B0G8M7 A0A1B0DGC8 A0A336KEN9 A0A1L8EEM8 B4LBV9 A0A0K8TS39 B4J0H9 B4H5V2 A0A1L8EEY1 Q2LYS4 A0A1B6CNU0 A0A1B6H879 A0A1B6MJC6 A0A1S3CX50 A0A3B0KGR9 W8C243 A0A1L8E035
Pubmed
19121390
22651552
26354079
26227816
25542738
22118469
+ More
29403074 25474469 24330624 27129103 25401762 26823975 25244985 24438588 12364791 14747013 17210077 17244540 24945155 26483478 23691247 17510324 20920257 23761445 20566863 20798317 25315136 24508170 30249741 17994087 26108605 26369729 15632085 24495485
29403074 25474469 24330624 27129103 25401762 26823975 25244985 24438588 12364791 14747013 17210077 17244540 24945155 26483478 23691247 17510324 20920257 23761445 20566863 20798317 25315136 24508170 30249741 17994087 26108605 26369729 15632085 24495485
EMBL
BABH01016398
DQ311424
ABD36368.1
AK401773
KQ459606
BAM18395.1
+ More
KPI91717.1 KQ460779 KPJ12303.1 JTDY01001566 KOB73510.1 RSAL01000007 RVE54102.1 NWSH01001104 PCG72597.1 KM358527 KM358528 AIW61836.1 AGBW02014084 OWR42292.1 ODYU01012166 SOQ58335.1 PYGN01000029 PSN56985.1 GFTR01002261 JAW14165.1 NEVH01003752 PNF40029.1 GBBI01000237 JAC18475.1 GEMB01002198 JAS00982.1 DS231867 EDS40660.1 GALA01001381 JAA93471.1 ACPB03012524 GDKW01003513 JAI53082.1 GBHO01004524 GBRD01001457 GDHC01009880 JAG39080.1 JAG64364.1 JAQ08749.1 AXCM01002771 GFDL01007530 JAV27515.1 ATLV01010511 KE524585 KFB35552.1 GGFK01004340 MBW37661.1 GGFK01008693 MBW42014.1 AAAB01008807 EAA04498.2 APCN01000178 GGFJ01010521 MBW59662.1 GGFJ01010503 MBW59644.1 GAMD01002865 JAA98725.1 JXJN01003556 GGFM01007435 MBW28186.1 AY826134 GAPW01005587 AAV90706.1 JAC08011.1 JXUM01047031 KQ561501 KXJ78378.1 GAPW01005588 JAC08010.1 AK417591 BAN20806.1 AXCN02000540 CH477576 EAT38787.1 GGFM01004454 MBW25205.1 GGFM01003921 MBW24672.1 ADMH02001146 ETN63938.1 CVRI01000059 CRL03325.1 KK854505 PTY16427.1 DS235815 EEB17514.1 GL451480 EFN79238.1 KK107046 QOIP01000004 EZA61799.1 RLU23904.1 CH964062 EDW78803.1 JRES01001499 KNC22421.1 AJVK01030297 CH902618 EDV38691.1 CCAG010000290 AJVK01059527 UFQS01000211 UFQT01000211 SSX01363.1 SSX21743.1 GFDG01001651 JAV17148.1 CH940647 EDW69759.1 GDAI01000436 JAI17167.1 CH916366 EDV96815.1 CH479212 EDW33169.1 GFDG01001650 JAV17149.1 CH379069 EAL29786.2 GEDC01022245 JAS15053.1 GECU01036789 JAS70917.1 GEBQ01030180 GEBQ01003966 JAT09797.1 JAT36011.1 OUUW01000012 SPP87620.1 GAMC01000653 JAC05903.1 GFDF01001964 JAV12120.1
KPI91717.1 KQ460779 KPJ12303.1 JTDY01001566 KOB73510.1 RSAL01000007 RVE54102.1 NWSH01001104 PCG72597.1 KM358527 KM358528 AIW61836.1 AGBW02014084 OWR42292.1 ODYU01012166 SOQ58335.1 PYGN01000029 PSN56985.1 GFTR01002261 JAW14165.1 NEVH01003752 PNF40029.1 GBBI01000237 JAC18475.1 GEMB01002198 JAS00982.1 DS231867 EDS40660.1 GALA01001381 JAA93471.1 ACPB03012524 GDKW01003513 JAI53082.1 GBHO01004524 GBRD01001457 GDHC01009880 JAG39080.1 JAG64364.1 JAQ08749.1 AXCM01002771 GFDL01007530 JAV27515.1 ATLV01010511 KE524585 KFB35552.1 GGFK01004340 MBW37661.1 GGFK01008693 MBW42014.1 AAAB01008807 EAA04498.2 APCN01000178 GGFJ01010521 MBW59662.1 GGFJ01010503 MBW59644.1 GAMD01002865 JAA98725.1 JXJN01003556 GGFM01007435 MBW28186.1 AY826134 GAPW01005587 AAV90706.1 JAC08011.1 JXUM01047031 KQ561501 KXJ78378.1 GAPW01005588 JAC08010.1 AK417591 BAN20806.1 AXCN02000540 CH477576 EAT38787.1 GGFM01004454 MBW25205.1 GGFM01003921 MBW24672.1 ADMH02001146 ETN63938.1 CVRI01000059 CRL03325.1 KK854505 PTY16427.1 DS235815 EEB17514.1 GL451480 EFN79238.1 KK107046 QOIP01000004 EZA61799.1 RLU23904.1 CH964062 EDW78803.1 JRES01001499 KNC22421.1 AJVK01030297 CH902618 EDV38691.1 CCAG010000290 AJVK01059527 UFQS01000211 UFQT01000211 SSX01363.1 SSX21743.1 GFDG01001651 JAV17148.1 CH940647 EDW69759.1 GDAI01000436 JAI17167.1 CH916366 EDV96815.1 CH479212 EDW33169.1 GFDG01001650 JAV17149.1 CH379069 EAL29786.2 GEDC01022245 JAS15053.1 GECU01036789 JAS70917.1 GEBQ01030180 GEBQ01003966 JAT09797.1 JAT36011.1 OUUW01000012 SPP87620.1 GAMC01000653 JAC05903.1 GFDF01001964 JAV12120.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000283053
UP000218220
+ More
UP000007151 UP000245037 UP000235965 UP000002320 UP000015103 UP000075883 UP000076408 UP000075920 UP000030765 UP000092445 UP000075902 UP000075885 UP000075903 UP000076407 UP000007062 UP000075840 UP000075884 UP000075881 UP000069272 UP000092460 UP000069940 UP000249989 UP000075886 UP000008820 UP000075900 UP000075880 UP000000673 UP000183832 UP000078200 UP000009046 UP000008237 UP000095301 UP000053097 UP000279307 UP000007798 UP000095300 UP000037069 UP000092462 UP000192221 UP000007801 UP000092444 UP000008792 UP000001070 UP000008744 UP000001819 UP000079169 UP000268350
UP000007151 UP000245037 UP000235965 UP000002320 UP000015103 UP000075883 UP000076408 UP000075920 UP000030765 UP000092445 UP000075902 UP000075885 UP000075903 UP000076407 UP000007062 UP000075840 UP000075884 UP000075881 UP000069272 UP000092460 UP000069940 UP000249989 UP000075886 UP000008820 UP000075900 UP000075880 UP000000673 UP000183832 UP000078200 UP000009046 UP000008237 UP000095301 UP000053097 UP000279307 UP000007798 UP000095300 UP000037069 UP000092462 UP000192221 UP000007801 UP000092444 UP000008792 UP000001070 UP000008744 UP000001819 UP000079169 UP000268350
PRIDE
Interpro
IPR013766
Thioredoxin_domain
+ More
IPR005746 Thioredoxin
IPR036249 Thioredoxin-like_sf
IPR017937 Thioredoxin_CS
IPR019327 DUF2373
IPR016024 ARM-type_fold
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR003891 Initiation_fac_eIF4g_MI
IPR003890 MIF4G-like_typ-3
IPR016021 MIF4-like_sf
IPR005746 Thioredoxin
IPR036249 Thioredoxin-like_sf
IPR017937 Thioredoxin_CS
IPR019327 DUF2373
IPR016024 ARM-type_fold
IPR001753 Enoyl-CoA_hydra/iso
IPR029045 ClpP/crotonase-like_dom_sf
IPR003891 Initiation_fac_eIF4g_MI
IPR003890 MIF4G-like_typ-3
IPR016021 MIF4-like_sf
Gene 3D
ProteinModelPortal
Q2F5J9
I4DKF5
A0A194R3C0
A0A0L7LDN2
A0A3S2M9J8
A0A2A4JKL4
+ More
A0A0A0VA40 A0A212ELD1 A0A2H1WZ26 A0A2P8ZKG6 A0A224XNX0 A0A2J7RGR7 A0A023FAV9 A0A161N1J1 B0WA02 T1DH89 T1HM49 A0A0P4VHF6 A0A0A9ZB97 A0A182MSJ7 A0A182YMK2 A0A1Q3FJ16 A0A182WJS9 A0A084VC58 A0A2M4AA76 A0A2M4AMK8 A0A1A9Z673 A0A182UA47 A0A182PDN6 A0A182VF20 A0A182XLI1 Q7QJ17 A0A182I2P7 A0A182NJY5 A0A182K161 A0A2M4C2Y0 A0A182FGK8 A0A2M4C2X2 T1DNH7 A0A1B0AU90 A0A2M3ZI13 Q5MIS6 A0A182G788 A0A023EGR0 R4WPK4 A0A182QER1 Q16W35 A0A2M3Z9Q5 A0A2M3Z830 A0A182R3K2 A0A182J5U0 W5JKW7 A0A1J1ISX0 A0A2R7W9U2 A0A1A9VTX9 E0VVV8 E2BYJ1 A0A1I8NEF7 A0A026X324 B4N364 A0A1I8PYY6 A0A0L0BQY9 A0A1B0DBV4 A0A1W4W3Q4 B3M6P1 A0A1B0G8M7 A0A1B0DGC8 A0A336KEN9 A0A1L8EEM8 B4LBV9 A0A0K8TS39 B4J0H9 B4H5V2 A0A1L8EEY1 Q2LYS4 A0A1B6CNU0 A0A1B6H879 A0A1B6MJC6 A0A1S3CX50 A0A3B0KGR9 W8C243 A0A1L8E035
A0A0A0VA40 A0A212ELD1 A0A2H1WZ26 A0A2P8ZKG6 A0A224XNX0 A0A2J7RGR7 A0A023FAV9 A0A161N1J1 B0WA02 T1DH89 T1HM49 A0A0P4VHF6 A0A0A9ZB97 A0A182MSJ7 A0A182YMK2 A0A1Q3FJ16 A0A182WJS9 A0A084VC58 A0A2M4AA76 A0A2M4AMK8 A0A1A9Z673 A0A182UA47 A0A182PDN6 A0A182VF20 A0A182XLI1 Q7QJ17 A0A182I2P7 A0A182NJY5 A0A182K161 A0A2M4C2Y0 A0A182FGK8 A0A2M4C2X2 T1DNH7 A0A1B0AU90 A0A2M3ZI13 Q5MIS6 A0A182G788 A0A023EGR0 R4WPK4 A0A182QER1 Q16W35 A0A2M3Z9Q5 A0A2M3Z830 A0A182R3K2 A0A182J5U0 W5JKW7 A0A1J1ISX0 A0A2R7W9U2 A0A1A9VTX9 E0VVV8 E2BYJ1 A0A1I8NEF7 A0A026X324 B4N364 A0A1I8PYY6 A0A0L0BQY9 A0A1B0DBV4 A0A1W4W3Q4 B3M6P1 A0A1B0G8M7 A0A1B0DGC8 A0A336KEN9 A0A1L8EEM8 B4LBV9 A0A0K8TS39 B4J0H9 B4H5V2 A0A1L8EEY1 Q2LYS4 A0A1B6CNU0 A0A1B6H879 A0A1B6MJC6 A0A1S3CX50 A0A3B0KGR9 W8C243 A0A1L8E035
PDB
1W89
E-value=1.1679e-29,
Score=317
Ontologies
GO
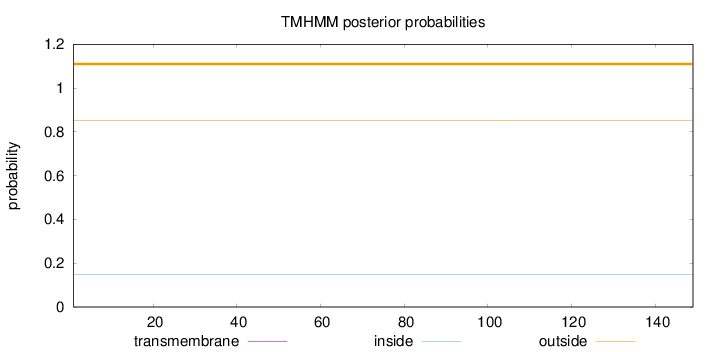
Topology
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00246
Exp number, first 60 AAs:
0.00153
Total prob of N-in:
0.14911
outside
1 - 149
Population Genetic Test Statistics
Pi
220.639584
Theta
195.639173
Tajima's D
0.438454
CLR
56.658091
CSRT
0.500674966251687
Interpretation
Uncertain
