Gene
KWMTBOMO06759
Pre Gene Modal
BGIBMGA012028
Annotation
PREDICTED:_pentatricopeptide_repeat-containing_protein_1?_mitochondrial_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 4.018
Sequence
CDS
ATGGCATGTACTAATCTGCTGAGAAATATTAAAACAATACATTTTATTCGACGTAGCTTTTTCTCAACTTCGATTATACAAAATAATCAATCTAAATATCCCAAACATACAGAATTCCGAACAGAACACAATCAATTGGTGTATTTAGATGATCCTGATAAGTTTGGGACATTGTCCGGTTTCAAAATACAGGACGAAAAACTTGTAGATGCTGGAGATATAAAAGAAGAAGAATATACACAAAATAAACCCTTACCTGGTAAAAAATTAACCACTAGACAGTATGCTGATTTAATTAAACAATACATAAAGCATAAAAGATTAAAAGAAGCTATTGATGTATTGGAGGTGAGAATGCTACAGGAAGATAAAGTGAAACCGGAAAATTATATATTCAACATATTAATTGGTGCGTGTGCTGATGTAGGTTACACAAAAAAAGCATTTCGTTTGTTCAATGAAATGAAAGCACGAGGACTAAAACCTACAGGTGATACTTATACCTGTTTGTTTCATGCATGTAGCAAAAGTCCATGGGCATCGGATGGTTTAAAAAACGCTAAACATCTTCGAGAACATATGATTTTAAAAGGAATAGAACCAAATCTGACGAATTATAATGCAATGATAAAGGCATTTGGATGTTGTGGTGATTTGGTTACGGCTTTCAGAATAGTTGATGAAATGGTAACTAAGAAAATAAAAATTAGAGTTCATACTTTTAACCATCTGCTACAAGCTTGTATATCAGATAAAAATAATGGACTGAGGCATGCTCTGATTGTATGGAGAAAAATGTTGAATGTTAGAGAGAAACCTAATATTTACTCTTTCAATTTAATGCTTAAATGTGTGAAAGACTGCAATTTGGGAACTAAAGAGGACATTGAAGAATTAGTTGGTGCTGTCAAAGGAAAAACATTAAAAGGAAGCAAAAATGATATGCTAGAGACGCAAAATATTGACTCGCAAGAGTTAAAATTATTGCAAGATCCTAAAAGTCCTCCTGAAAAAATTAGAAAAGACCATATTGATTCTCAAAAAAGTATTAATGGTAGTAACAAAATGAATATGCAAAATTCAGATGCAGTAGAAACAACAGATTTAATGATTACAGAAAACAAAGCAAAGATAACAATACAATTAGTAGATAATTTAAAAAATACCAGCAAAGAACAAAGAAATGTACCTAATTTATTGTCAAAGGAAATTAATATGCACCATATATTAGCTTTACAAGAAGTGCATTCATTACAAGATAAGTTTACTGTAATAGGTGGACAAGAAGATTTCCTTGAAGAAATGAGTGCTTATTCAGTTAAACCAGACATTAAAACGTTTACACAAATGCTCCCATTATTGGATAATACAACAGAGGCTGAAAATAAACTACTTCATACTATGCACAATATGAAGATACGAGCAGATATTGACTTCTACAACATGTTAATAAAGAAAAGGTGTTTAAGGCTGGATTATGATGCTGCTAAAGCTGTTAAAAAGATAATAAAAAAGGAAAGTGAAATTCGGAAACGTTTGTATCCAGATAATAAAAAGAAGAGACTAAATCTTAATATAATGTCTTATGGCGTCTTGGCAATGGCTTGTAAAACTAAAGAAGATGCAGAAATCCTCCTAGCTGATATGAGACAAGAACAACTTAAGGTGAATATAGAAATTCTTGGAGCTCTATTGAAACATGGCATCGTAAATTATCGATTCGACTACGTTCTGTACGTATTGAATATTGTAAAAGACGAAAAGGTTCCTGTTAGTGATTTGTTTTTAAAGAATTTAGAGGACTTCAATGAAAAATGTATGAAAATTATACAGCAGAATGAAAACAGTCATCGCAAATATTCCAATTCTTTCGAAACAGCTTACAATAAATTCTCGAAGATTTATACTAATTGGTTAAGAGAAGTAAACATTCATGAAGCTTTGAAGCAAGAACATCCTTGGGAACAATTTAAGACTTCCAGCCCAGAAACAGTGCAAAGGCAAAATATTAAAATCATCGAACCAAAGAGATTTAACAAAAGAAATCGTAAATATGTACCATATGTTCCTAAGATATAA
Protein
MACTNLLRNIKTIHFIRRSFFSTSIIQNNQSKYPKHTEFRTEHNQLVYLDDPDKFGTLSGFKIQDEKLVDAGDIKEEEYTQNKPLPGKKLTTRQYADLIKQYIKHKRLKEAIDVLEVRMLQEDKVKPENYIFNILIGACADVGYTKKAFRLFNEMKARGLKPTGDTYTCLFHACSKSPWASDGLKNAKHLREHMILKGIEPNLTNYNAMIKAFGCCGDLVTAFRIVDEMVTKKIKIRVHTFNHLLQACISDKNNGLRHALIVWRKMLNVREKPNIYSFNLMLKCVKDCNLGTKEDIEELVGAVKGKTLKGSKNDMLETQNIDSQELKLLQDPKSPPEKIRKDHIDSQKSINGSNKMNMQNSDAVETTDLMITENKAKITIQLVDNLKNTSKEQRNVPNLLSKEINMHHILALQEVHSLQDKFTVIGGQEDFLEEMSAYSVKPDIKTFTQMLPLLDNTTEAENKLLHTMHNMKIRADIDFYNMLIKKRCLRLDYDAAKAVKKIIKKESEIRKRLYPDNKKKRLNLNIMSYGVLAMACKTKEDAEILLADMRQEQLKVNIEILGALLKHGIVNYRFDYVLYVLNIVKDEKVPVSDLFLKNLEDFNEKCMKIIQQNENSHRKYSNSFETAYNKFSKIYTNWLREVNIHEALKQEHPWEQFKTSSPETVQRQNIKIIEPKRFNKRNRKYVPYVPKI
Summary
Uniprot
A0A2A4JMH5
A0A2H1WZ37
A0A212ELB0
A0A194R8V0
A0A194PKT8
A0A0L7LE76
+ More
A0A1S4FCT3 Q176Y9 A0A0T6B1Q6 A0A182GXS9 A0A182NKG0 A0A182G5T5 A0A182R726 A0A182Q6L1 A0A182JX20 B0VZJ7 U5EGC6 A0A182PJT2 A0A182SG24 A0A084W446 A0A182XM40 A0A182MB69 A0A182VEF3 A0A182KPA7 A0A182I1M4 A0A182YR13 Q7Q645 A0A182UJB5 A0A182IQQ0 A0A182WHN6 A0A1J1J4U0 A0A1B0CB15 A0A182FD49 A0A2J7QW91 A0A1W4WRQ8 D6WG40 A0A1Y1K6Z0 A0A1B6FRY6 B4IWZ9 A0A336K072 A0A1B0CYP6 B4KWZ3 A0A2H8TDJ6 Q29FC7 B4H1I3 A0A1I8QCC8 A0A0A1WPX3 A0A2M4APC2 A0A2M4APC0 U4U9Z0 A0A2M4APG0 A0A3B0KTE5 A0A1A9VN20 B4LFQ0 W5JIQ6 A0A1A9X0I1 A0A1W4URV1 A0A1I8MYB1 A0A1A9ZMI7 B4HUD5 N6U5U9 A0A0K8W507 A0A1B0AQD1 A0A1A9XIM0 A0A1B0GGK1 B3M406 W8BSS3 B4MXF4 A0A0J9RQR6 B4QRI2 Q9VRJ7 B4PIE1 B3NG92 A0A2T7Q0Z1 A0A146LMM2 A0A146LV43 A0A0P5ED92 A0A023F079 T1HPW7 A0A0N8EB12 A0A0P6F6R5 A0A0V0G849 A0A0P5UDL2 A0A0N8BEL4 A0A0P4ZMD3 A0A0P5IF28 A0A0P5DDN6 A0A0P5JDR9 A0A0P5SKX6 A0A0P5TR25 A0A0N8CUR3 A0A0P5T0C0 A0A0P5DM19 A0A0P4ZSQ7 A0A0P5RU21 A0A0P5NC12 A0A3R7M0S5
A0A1S4FCT3 Q176Y9 A0A0T6B1Q6 A0A182GXS9 A0A182NKG0 A0A182G5T5 A0A182R726 A0A182Q6L1 A0A182JX20 B0VZJ7 U5EGC6 A0A182PJT2 A0A182SG24 A0A084W446 A0A182XM40 A0A182MB69 A0A182VEF3 A0A182KPA7 A0A182I1M4 A0A182YR13 Q7Q645 A0A182UJB5 A0A182IQQ0 A0A182WHN6 A0A1J1J4U0 A0A1B0CB15 A0A182FD49 A0A2J7QW91 A0A1W4WRQ8 D6WG40 A0A1Y1K6Z0 A0A1B6FRY6 B4IWZ9 A0A336K072 A0A1B0CYP6 B4KWZ3 A0A2H8TDJ6 Q29FC7 B4H1I3 A0A1I8QCC8 A0A0A1WPX3 A0A2M4APC2 A0A2M4APC0 U4U9Z0 A0A2M4APG0 A0A3B0KTE5 A0A1A9VN20 B4LFQ0 W5JIQ6 A0A1A9X0I1 A0A1W4URV1 A0A1I8MYB1 A0A1A9ZMI7 B4HUD5 N6U5U9 A0A0K8W507 A0A1B0AQD1 A0A1A9XIM0 A0A1B0GGK1 B3M406 W8BSS3 B4MXF4 A0A0J9RQR6 B4QRI2 Q9VRJ7 B4PIE1 B3NG92 A0A2T7Q0Z1 A0A146LMM2 A0A146LV43 A0A0P5ED92 A0A023F079 T1HPW7 A0A0N8EB12 A0A0P6F6R5 A0A0V0G849 A0A0P5UDL2 A0A0N8BEL4 A0A0P4ZMD3 A0A0P5IF28 A0A0P5DDN6 A0A0P5JDR9 A0A0P5SKX6 A0A0P5TR25 A0A0N8CUR3 A0A0P5T0C0 A0A0P5DM19 A0A0P4ZSQ7 A0A0P5RU21 A0A0P5NC12 A0A3R7M0S5
Pubmed
22118469
26354079
26227816
17510324
26483478
24438588
+ More
20966253 25244985 12364791 14747013 17210077 18362917 19820115 28004739 17994087 15632085 25830018 23537049 20920257 23761445 25315136 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26823975 25474469
20966253 25244985 12364791 14747013 17210077 18362917 19820115 28004739 17994087 15632085 25830018 23537049 20920257 23761445 25315136 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26823975 25474469
EMBL
NWSH01001104
PCG72600.1
ODYU01012166
SOQ58333.1
AGBW02014084
OWR42290.1
+ More
KQ460779 KPJ12301.1 KQ459606 KPI91715.1 JTDY01001566 KOB73511.1 CH477380 EAT42220.1 LJIG01016199 KRT81318.1 JXUM01020768 KQ560581 KXJ81811.1 JXUM01043912 KQ561380 KXJ78776.1 AXCN02000399 DS231814 EDS31734.1 GANO01003498 JAB56373.1 ATLV01020275 KE525297 KFB44990.1 AXCM01004514 APCN01000069 AAAB01008960 EAA11660.4 CVRI01000069 CRL06978.1 AJWK01004728 NEVH01009766 PNF32841.1 KQ971319 EFA00524.1 GEZM01090985 JAV57144.1 GECZ01016819 JAS52950.1 CH916366 EDV96305.1 UFQS01000040 UFQT01000040 SSW98139.1 SSX18525.1 AJVK01020262 AJVK01020263 AJVK01020264 CH933809 EDW18614.2 GFXV01000354 MBW12159.1 CH379067 EAL31328.1 CH479202 EDW30160.1 GBXI01013586 JAD00706.1 GGFK01009299 MBW42620.1 GGFK01009137 MBW42458.1 KB632256 ERL90759.1 GGFK01009338 MBW42659.1 OUUW01000012 SPP87168.1 CH940647 EDW69277.1 ADMH02001390 ETN62679.1 CH480817 EDW50556.1 APGK01041328 KB740993 ENN76016.1 GDHF01006172 JAI46142.1 JXJN01001810 CCAG010020395 CH902618 EDV40368.1 GAMC01002135 JAC04421.1 CH963876 EDW76723.1 CM002912 KMY97704.1 CM000363 EDX09293.1 AE014296 BT011031 AAF50798.1 AAR30191.1 CM000159 EDW93483.1 CH954178 EDV50993.1 PZQS01000001 PVD39344.1 GDHC01010989 JAQ07640.1 GDHC01007674 JAQ10955.1 GDIP01143078 JAJ80324.1 GBBI01003819 JAC14893.1 ACPB03001903 GDIQ01044372 JAN50365.1 GDIQ01052562 JAN42175.1 GECL01001833 JAP04291.1 GDIP01131748 JAL71966.1 GDIQ01185347 JAK66378.1 GDIP01211275 JAJ12127.1 GDIQ01222049 JAK29676.1 GDIP01162935 JAJ60467.1 GDIQ01201001 JAK50724.1 GDIP01138489 JAL65225.1 GDIP01140736 JAL62978.1 GDIP01096943 JAM06772.1 GDIP01139594 JAL64120.1 GDIP01157736 JAJ65666.1 GDIP01210127 JAJ13275.1 GDIQ01097140 JAL54586.1 GDIQ01147370 JAL04356.1 QCYY01002655 ROT68584.1
KQ460779 KPJ12301.1 KQ459606 KPI91715.1 JTDY01001566 KOB73511.1 CH477380 EAT42220.1 LJIG01016199 KRT81318.1 JXUM01020768 KQ560581 KXJ81811.1 JXUM01043912 KQ561380 KXJ78776.1 AXCN02000399 DS231814 EDS31734.1 GANO01003498 JAB56373.1 ATLV01020275 KE525297 KFB44990.1 AXCM01004514 APCN01000069 AAAB01008960 EAA11660.4 CVRI01000069 CRL06978.1 AJWK01004728 NEVH01009766 PNF32841.1 KQ971319 EFA00524.1 GEZM01090985 JAV57144.1 GECZ01016819 JAS52950.1 CH916366 EDV96305.1 UFQS01000040 UFQT01000040 SSW98139.1 SSX18525.1 AJVK01020262 AJVK01020263 AJVK01020264 CH933809 EDW18614.2 GFXV01000354 MBW12159.1 CH379067 EAL31328.1 CH479202 EDW30160.1 GBXI01013586 JAD00706.1 GGFK01009299 MBW42620.1 GGFK01009137 MBW42458.1 KB632256 ERL90759.1 GGFK01009338 MBW42659.1 OUUW01000012 SPP87168.1 CH940647 EDW69277.1 ADMH02001390 ETN62679.1 CH480817 EDW50556.1 APGK01041328 KB740993 ENN76016.1 GDHF01006172 JAI46142.1 JXJN01001810 CCAG010020395 CH902618 EDV40368.1 GAMC01002135 JAC04421.1 CH963876 EDW76723.1 CM002912 KMY97704.1 CM000363 EDX09293.1 AE014296 BT011031 AAF50798.1 AAR30191.1 CM000159 EDW93483.1 CH954178 EDV50993.1 PZQS01000001 PVD39344.1 GDHC01010989 JAQ07640.1 GDHC01007674 JAQ10955.1 GDIP01143078 JAJ80324.1 GBBI01003819 JAC14893.1 ACPB03001903 GDIQ01044372 JAN50365.1 GDIQ01052562 JAN42175.1 GECL01001833 JAP04291.1 GDIP01131748 JAL71966.1 GDIQ01185347 JAK66378.1 GDIP01211275 JAJ12127.1 GDIQ01222049 JAK29676.1 GDIP01162935 JAJ60467.1 GDIQ01201001 JAK50724.1 GDIP01138489 JAL65225.1 GDIP01140736 JAL62978.1 GDIP01096943 JAM06772.1 GDIP01139594 JAL64120.1 GDIP01157736 JAJ65666.1 GDIP01210127 JAJ13275.1 GDIQ01097140 JAL54586.1 GDIQ01147370 JAL04356.1 QCYY01002655 ROT68584.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
UP000008820
+ More
UP000069940 UP000249989 UP000075884 UP000075900 UP000075886 UP000075881 UP000002320 UP000075885 UP000075901 UP000030765 UP000076407 UP000075883 UP000075903 UP000075882 UP000075840 UP000076408 UP000007062 UP000075902 UP000075880 UP000075920 UP000183832 UP000092461 UP000069272 UP000235965 UP000192223 UP000007266 UP000001070 UP000092462 UP000009192 UP000001819 UP000008744 UP000095300 UP000030742 UP000268350 UP000078200 UP000008792 UP000000673 UP000091820 UP000192221 UP000095301 UP000092445 UP000001292 UP000019118 UP000092460 UP000092443 UP000092444 UP000007801 UP000007798 UP000000304 UP000000803 UP000002282 UP000008711 UP000245119 UP000015103 UP000283509
UP000069940 UP000249989 UP000075884 UP000075900 UP000075886 UP000075881 UP000002320 UP000075885 UP000075901 UP000030765 UP000076407 UP000075883 UP000075903 UP000075882 UP000075840 UP000076408 UP000007062 UP000075902 UP000075880 UP000075920 UP000183832 UP000092461 UP000069272 UP000235965 UP000192223 UP000007266 UP000001070 UP000092462 UP000009192 UP000001819 UP000008744 UP000095300 UP000030742 UP000268350 UP000078200 UP000008792 UP000000673 UP000091820 UP000192221 UP000095301 UP000092445 UP000001292 UP000019118 UP000092460 UP000092443 UP000092444 UP000007801 UP000007798 UP000000304 UP000000803 UP000002282 UP000008711 UP000245119 UP000015103 UP000283509
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
Gene 3D
ProteinModelPortal
A0A2A4JMH5
A0A2H1WZ37
A0A212ELB0
A0A194R8V0
A0A194PKT8
A0A0L7LE76
+ More
A0A1S4FCT3 Q176Y9 A0A0T6B1Q6 A0A182GXS9 A0A182NKG0 A0A182G5T5 A0A182R726 A0A182Q6L1 A0A182JX20 B0VZJ7 U5EGC6 A0A182PJT2 A0A182SG24 A0A084W446 A0A182XM40 A0A182MB69 A0A182VEF3 A0A182KPA7 A0A182I1M4 A0A182YR13 Q7Q645 A0A182UJB5 A0A182IQQ0 A0A182WHN6 A0A1J1J4U0 A0A1B0CB15 A0A182FD49 A0A2J7QW91 A0A1W4WRQ8 D6WG40 A0A1Y1K6Z0 A0A1B6FRY6 B4IWZ9 A0A336K072 A0A1B0CYP6 B4KWZ3 A0A2H8TDJ6 Q29FC7 B4H1I3 A0A1I8QCC8 A0A0A1WPX3 A0A2M4APC2 A0A2M4APC0 U4U9Z0 A0A2M4APG0 A0A3B0KTE5 A0A1A9VN20 B4LFQ0 W5JIQ6 A0A1A9X0I1 A0A1W4URV1 A0A1I8MYB1 A0A1A9ZMI7 B4HUD5 N6U5U9 A0A0K8W507 A0A1B0AQD1 A0A1A9XIM0 A0A1B0GGK1 B3M406 W8BSS3 B4MXF4 A0A0J9RQR6 B4QRI2 Q9VRJ7 B4PIE1 B3NG92 A0A2T7Q0Z1 A0A146LMM2 A0A146LV43 A0A0P5ED92 A0A023F079 T1HPW7 A0A0N8EB12 A0A0P6F6R5 A0A0V0G849 A0A0P5UDL2 A0A0N8BEL4 A0A0P4ZMD3 A0A0P5IF28 A0A0P5DDN6 A0A0P5JDR9 A0A0P5SKX6 A0A0P5TR25 A0A0N8CUR3 A0A0P5T0C0 A0A0P5DM19 A0A0P4ZSQ7 A0A0P5RU21 A0A0P5NC12 A0A3R7M0S5
A0A1S4FCT3 Q176Y9 A0A0T6B1Q6 A0A182GXS9 A0A182NKG0 A0A182G5T5 A0A182R726 A0A182Q6L1 A0A182JX20 B0VZJ7 U5EGC6 A0A182PJT2 A0A182SG24 A0A084W446 A0A182XM40 A0A182MB69 A0A182VEF3 A0A182KPA7 A0A182I1M4 A0A182YR13 Q7Q645 A0A182UJB5 A0A182IQQ0 A0A182WHN6 A0A1J1J4U0 A0A1B0CB15 A0A182FD49 A0A2J7QW91 A0A1W4WRQ8 D6WG40 A0A1Y1K6Z0 A0A1B6FRY6 B4IWZ9 A0A336K072 A0A1B0CYP6 B4KWZ3 A0A2H8TDJ6 Q29FC7 B4H1I3 A0A1I8QCC8 A0A0A1WPX3 A0A2M4APC2 A0A2M4APC0 U4U9Z0 A0A2M4APG0 A0A3B0KTE5 A0A1A9VN20 B4LFQ0 W5JIQ6 A0A1A9X0I1 A0A1W4URV1 A0A1I8MYB1 A0A1A9ZMI7 B4HUD5 N6U5U9 A0A0K8W507 A0A1B0AQD1 A0A1A9XIM0 A0A1B0GGK1 B3M406 W8BSS3 B4MXF4 A0A0J9RQR6 B4QRI2 Q9VRJ7 B4PIE1 B3NG92 A0A2T7Q0Z1 A0A146LMM2 A0A146LV43 A0A0P5ED92 A0A023F079 T1HPW7 A0A0N8EB12 A0A0P6F6R5 A0A0V0G849 A0A0P5UDL2 A0A0N8BEL4 A0A0P4ZMD3 A0A0P5IF28 A0A0P5DDN6 A0A0P5JDR9 A0A0P5SKX6 A0A0P5TR25 A0A0N8CUR3 A0A0P5T0C0 A0A0P5DM19 A0A0P4ZSQ7 A0A0P5RU21 A0A0P5NC12 A0A3R7M0S5
PDB
5I9G
E-value=6.26063e-17,
Score=216
Ontologies
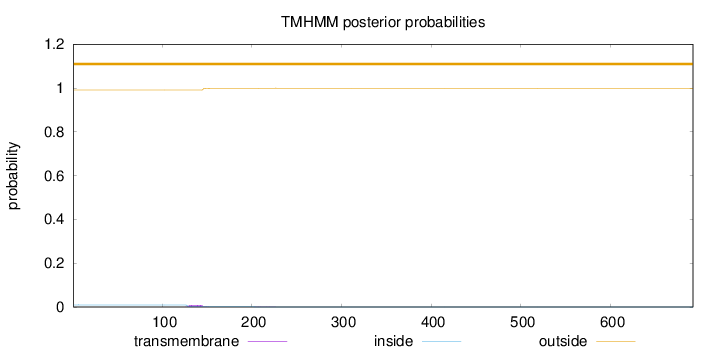
Topology
Length:
692
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17116
Exp number, first 60 AAs:
0.00093
Total prob of N-in:
0.00948
outside
1 - 692
Population Genetic Test Statistics
Pi
132.521658
Theta
145.618358
Tajima's D
-0.647463
CLR
1.073967
CSRT
0.214789260536973
Interpretation
Uncertain
