Pre Gene Modal
BGIBMGA012127
Annotation
PREDICTED:_glutamate-rich_WD_repeat-containing_protein_1_[Amyelois_transitella]
Full name
Anoctamin
Location in the cell
Nuclear Reliability : 3.986
Sequence
CDS
ATGGACTCAAGTTCAGAGAATGAAGACGACGAAATGAATGAAGAAGCGGAGAGTGACCAGAACGATGATCGCCCGAAAACTTATTTACCAGACCAGCCTTTAAAGGAAGATGAACAATTGGTCTGTGATCATTCTGCTTATGTGATGCTGCATCAGGCTCAAACTGGAGCCCCATGTCTTAGCTTTGACATAGTTACAGACAATCTTGGAAATGATCGAAATGAATTTCCTATGACTGCTTATTTGGTCGCCGGAACACAAGCGTCAAGTGCACATTTAAATAATCTTTTGGTAATAAAAATGTCAAACCTACACCCTATATCTAAACCAGAGGAAGATGAAGAATCTGAAGATGATGATTCTGATGATGAAGATGAAGAGCAGAAACCACAGATGACATTCTCATTCATAAAACATCAAGGATGTGTAAACAGAATAAGGGCTACAAATTTTAAAAACTCTGTATTGGCAGCAAGTTGGTCAGAACTAGGCCGAGTTGACATTTGGAATATCACACAACAGCTGCAAGCCGTGGATGACCCAGTCGTGTTAGAAAGATACAACTTGGAGACAGTTTCAAATCCCGTCAAACCTATATACTCATTTAACGGGCATCAGCAAGAGGGTTTCGCTCTCGATTGGTGTCCTACTGAATCAGGTATGCTGGCAACTGGAGACTGCAGAAGAGACATTCATATATGGAGACCAAATGAAGGTGGCACTTGGACTGTAGACCAACGACCCCTTGTAGGGCATACCAGTTCTGTTGAAGACATACAATGGTCTCCAAATGAAAGGAACGTATTAGCTACATGCTCTGTAGATAAGACCATAAGAATATGGGATACTAGAGCAGCACCACAAAAGGCTTGCATGCTCACAGCTGAAAATGCACATCAAAGTGACATTAATGTTATATCATGGAATGGAAAAGAGCCATTTATTGCAAGCGGCGGTGATGATGGTTTTCTACATATCTGGGACTTGAGACAGTTTACAAATAGTACTCCAGTGGGTACATTCAAACATCACACTGCACCAGTTACATCAGTGGAATGGCATTGGACCGAGCCCAGCGTGTTAGCATCAGCAGGAGAAGATAATCAAGTAGCTCTTTGGGACTTAGCGGTTGAAAAAGACGATGATGAAGTTGAAGATGATGATGAATTAAAGGTTTGTCTATTTATTAACGTAATCAATTAA
Protein
MDSSSENEDDEMNEEAESDQNDDRPKTYLPDQPLKEDEQLVCDHSAYVMLHQAQTGAPCLSFDIVTDNLGNDRNEFPMTAYLVAGTQASSAHLNNLLVIKMSNLHPISKPEEDEESEDDDSDDEDEEQKPQMTFSFIKHQGCVNRIRATNFKNSVLAASWSELGRVDIWNITQQLQAVDDPVVLERYNLETVSNPVKPIYSFNGHQQEGFALDWCPTESGMLATGDCRRDIHIWRPNEGGTWTVDQRPLVGHTSSVEDIQWSPNERNVLATCSVDKTIRIWDTRAAPQKACMLTAENAHQSDINVISWNGKEPFIASGGDDGFLHIWDLRQFTNSTPVGTFKHHTAPVTSVEWHWTEPSVLASAGEDNQVALWDLAVEKDDDEVEDDDELKVCLFINVIN
Summary
Similarity
Belongs to the 14-3-3 family.
Belongs to the anoctamin family.
Belongs to the anoctamin family.
Uniprot
H9JRG7
A0A212ELD8
A0A2H1WZA0
A0A3S2TSQ5
A0A2A4JKW4
A0A194R4Y3
+ More
A0A194PG85 D6WH34 B4KM61 B4LNM8 A0A1I8PI13 A0A0M4E7E4 A0A1A9YE60 A0A0M4E580 B4J5Q6 A0A1B0FG95 B4H8C7 A0A1A9ZCA6 Q28YP6 A0A3B0JXJ6 A0A0L0CBR3 A0A1A9VQJ2 A0A1I8MWQ7 T1P9E4 B4II06 B4QCP0 B3N3I8 A0A182YEL3 T1DM33 A0A1A9WWK2 A0A034VFS4 A0A1Q3EXF6 B4NYX9 A0A1L8DX10 A0A1W4WA68 A0A182WLI6 A0A0K8VTZ3 A0A182M8R8 Q8SYL1 A0A2M4BN27 A0A2M4BMA6 A0A182FMQ9 A0A2M4AEI2 W5J9U4 W8B227 A0A2M4AE82 A0A2M4AE84 A0A182P0N6 A0A2M3ZHX6 A0A182QIL7 A0A182JND6 A0A2M4BM72 A0A182SW66 Q16IG2 B4NMV5 A0A182NK56 A0A182RP63 A0A2M4BQ15 B0XBR5 A0A182WWZ4 A0A182UT22 A0A182TFD3 A0A182I2J1 Q7QIY0 Q16U37 A0A2J7RKS2 A0A067R1H4 A0A182WWZ2 A0A0K8TRP1 A0A1B0D5R1 B3MBQ2 A0A182IKF6 N6UDW6 A0A084WK73 A0A1L8DWP7 E0VGG3 A0A1Y1N8J2 A0A336MAG9 A0A1S3H902 A0A0P5ZMX4 A0A0P5S9D2 A0A1J1IKG9 A0A1J1IH46 A0A0P5VAU6 A0A0N8AP86 R7TDG9 K1QL27 A0A0J7NZ36 A0A1B6H2A8 A0A026VV92 A0A210QGC0 E2A728 A0A0P5ZAW2 A0A151X1Q0 E9IE28 A0A0P6B6T9 A0A2A3E0B9 K7J320
A0A194PG85 D6WH34 B4KM61 B4LNM8 A0A1I8PI13 A0A0M4E7E4 A0A1A9YE60 A0A0M4E580 B4J5Q6 A0A1B0FG95 B4H8C7 A0A1A9ZCA6 Q28YP6 A0A3B0JXJ6 A0A0L0CBR3 A0A1A9VQJ2 A0A1I8MWQ7 T1P9E4 B4II06 B4QCP0 B3N3I8 A0A182YEL3 T1DM33 A0A1A9WWK2 A0A034VFS4 A0A1Q3EXF6 B4NYX9 A0A1L8DX10 A0A1W4WA68 A0A182WLI6 A0A0K8VTZ3 A0A182M8R8 Q8SYL1 A0A2M4BN27 A0A2M4BMA6 A0A182FMQ9 A0A2M4AEI2 W5J9U4 W8B227 A0A2M4AE82 A0A2M4AE84 A0A182P0N6 A0A2M3ZHX6 A0A182QIL7 A0A182JND6 A0A2M4BM72 A0A182SW66 Q16IG2 B4NMV5 A0A182NK56 A0A182RP63 A0A2M4BQ15 B0XBR5 A0A182WWZ4 A0A182UT22 A0A182TFD3 A0A182I2J1 Q7QIY0 Q16U37 A0A2J7RKS2 A0A067R1H4 A0A182WWZ2 A0A0K8TRP1 A0A1B0D5R1 B3MBQ2 A0A182IKF6 N6UDW6 A0A084WK73 A0A1L8DWP7 E0VGG3 A0A1Y1N8J2 A0A336MAG9 A0A1S3H902 A0A0P5ZMX4 A0A0P5S9D2 A0A1J1IKG9 A0A1J1IH46 A0A0P5VAU6 A0A0N8AP86 R7TDG9 K1QL27 A0A0J7NZ36 A0A1B6H2A8 A0A026VV92 A0A210QGC0 E2A728 A0A0P5ZAW2 A0A151X1Q0 E9IE28 A0A0P6B6T9 A0A2A3E0B9 K7J320
Pubmed
19121390
22118469
26354079
18362917
19820115
17994087
+ More
15632085 26108605 25315136 22936249 25244985 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 24495485 17510324 12364791 14747013 17210077 24845553 26369729 23537049 24438588 20566863 28004739 23254933 22992520 24508170 30249741 28812685 20798317 21282665 20075255
15632085 26108605 25315136 22936249 25244985 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 24495485 17510324 12364791 14747013 17210077 24845553 26369729 23537049 24438588 20566863 28004739 23254933 22992520 24508170 30249741 28812685 20798317 21282665 20075255
EMBL
BABH01016400
BABH01016401
BABH01016402
AGBW02014084
OWR42289.1
ODYU01012166
+ More
SOQ58332.1 RSAL01000007 RVE54099.1 NWSH01001104 PCG72601.1 KQ460779 KPJ12300.1 KQ459606 KPI91714.1 KQ971321 EFA00626.1 CH933808 EDW08725.1 CH940648 EDW62208.2 CP012524 ALC40434.1 ALC41378.1 CH916367 EDW01832.1 CCAG010021922 CH479223 EDW34962.1 CM000071 EAL25919.1 OUUW01000001 SPP75808.1 JRES01000737 KNC28874.1 KA645347 AFP59976.1 CH480841 EDW49532.1 CM000362 CM002911 EDX05829.1 KMY91651.1 CH954177 EDV59870.1 GAMD01000111 JAB01480.1 GAKP01017613 JAC41339.1 GFDL01015050 JAV19995.1 CM000157 EDW89830.1 GFDF01003138 JAV10946.1 GDHF01009991 JAI42323.1 AXCM01005409 AE013599 AY071477 AY128462 AAF57313.3 AAL49099.1 AAM75055.1 AGB93248.1 GGFJ01005037 MBW54178.1 GGFJ01005021 MBW54162.1 GGFK01005801 MBW39122.1 ADMH02001744 ETN61232.1 GAMC01011355 JAB95200.1 GGFK01005775 MBW39096.1 GGFK01005776 MBW39097.1 GGFM01007416 MBW28167.1 AXCN02000429 GGFJ01005036 MBW54177.1 CH478080 EAT34047.1 CH964282 EDW85694.1 GGFJ01005870 MBW55011.1 DS232648 EDS44414.1 APCN01000164 AAAB01008807 EAA04110.2 CH477632 EAT38048.1 NEVH01002708 PNF41436.1 KK852815 KDR15821.1 GDAI01000576 JAI17027.1 AJVK01025533 AJVK01025534 AJVK01025535 CH902619 EDV38198.2 APGK01026644 KB740635 KB632195 ENN79900.1 ERL89957.1 ATLV01024097 KE525349 KFB50617.1 GFDF01003241 JAV10843.1 DS235142 EEB12469.1 GEZM01013378 JAV92606.1 UFQT01000305 SSX23038.1 GDIP01042146 JAM61569.1 GDIP01142996 LRGB01001361 JAL60718.1 KZS12453.1 CVRI01000051 CRK99574.1 CRK99573.1 GDIP01102937 JAM00778.1 GDIQ01256371 JAJ95353.1 AMQN01013747 AMQN01013748 KB310448 ELT91552.1 JH818095 EKC37492.1 LBMM01000724 KMQ97625.1 GECZ01000991 JAS68778.1 KK107796 QOIP01000013 EZA47652.1 RLU15583.1 NEDP02003775 OWF47796.1 GL437252 EFN70760.1 GDIP01059200 JAM44515.1 KQ982585 KYQ54319.1 GL762556 EFZ21202.1 GDIP01021170 JAM82545.1 KZ288488 PBC25223.1
SOQ58332.1 RSAL01000007 RVE54099.1 NWSH01001104 PCG72601.1 KQ460779 KPJ12300.1 KQ459606 KPI91714.1 KQ971321 EFA00626.1 CH933808 EDW08725.1 CH940648 EDW62208.2 CP012524 ALC40434.1 ALC41378.1 CH916367 EDW01832.1 CCAG010021922 CH479223 EDW34962.1 CM000071 EAL25919.1 OUUW01000001 SPP75808.1 JRES01000737 KNC28874.1 KA645347 AFP59976.1 CH480841 EDW49532.1 CM000362 CM002911 EDX05829.1 KMY91651.1 CH954177 EDV59870.1 GAMD01000111 JAB01480.1 GAKP01017613 JAC41339.1 GFDL01015050 JAV19995.1 CM000157 EDW89830.1 GFDF01003138 JAV10946.1 GDHF01009991 JAI42323.1 AXCM01005409 AE013599 AY071477 AY128462 AAF57313.3 AAL49099.1 AAM75055.1 AGB93248.1 GGFJ01005037 MBW54178.1 GGFJ01005021 MBW54162.1 GGFK01005801 MBW39122.1 ADMH02001744 ETN61232.1 GAMC01011355 JAB95200.1 GGFK01005775 MBW39096.1 GGFK01005776 MBW39097.1 GGFM01007416 MBW28167.1 AXCN02000429 GGFJ01005036 MBW54177.1 CH478080 EAT34047.1 CH964282 EDW85694.1 GGFJ01005870 MBW55011.1 DS232648 EDS44414.1 APCN01000164 AAAB01008807 EAA04110.2 CH477632 EAT38048.1 NEVH01002708 PNF41436.1 KK852815 KDR15821.1 GDAI01000576 JAI17027.1 AJVK01025533 AJVK01025534 AJVK01025535 CH902619 EDV38198.2 APGK01026644 KB740635 KB632195 ENN79900.1 ERL89957.1 ATLV01024097 KE525349 KFB50617.1 GFDF01003241 JAV10843.1 DS235142 EEB12469.1 GEZM01013378 JAV92606.1 UFQT01000305 SSX23038.1 GDIP01042146 JAM61569.1 GDIP01142996 LRGB01001361 JAL60718.1 KZS12453.1 CVRI01000051 CRK99574.1 CRK99573.1 GDIP01102937 JAM00778.1 GDIQ01256371 JAJ95353.1 AMQN01013747 AMQN01013748 KB310448 ELT91552.1 JH818095 EKC37492.1 LBMM01000724 KMQ97625.1 GECZ01000991 JAS68778.1 KK107796 QOIP01000013 EZA47652.1 RLU15583.1 NEDP02003775 OWF47796.1 GL437252 EFN70760.1 GDIP01059200 JAM44515.1 KQ982585 KYQ54319.1 GL762556 EFZ21202.1 GDIP01021170 JAM82545.1 KZ288488 PBC25223.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053240
UP000053268
+ More
UP000007266 UP000009192 UP000008792 UP000095300 UP000092553 UP000092443 UP000001070 UP000092444 UP000008744 UP000092445 UP000001819 UP000268350 UP000037069 UP000078200 UP000095301 UP000001292 UP000000304 UP000008711 UP000076408 UP000091820 UP000002282 UP000192221 UP000075920 UP000075883 UP000000803 UP000069272 UP000000673 UP000075885 UP000075886 UP000075881 UP000075901 UP000008820 UP000007798 UP000075884 UP000075900 UP000002320 UP000076407 UP000075903 UP000075902 UP000075840 UP000007062 UP000235965 UP000027135 UP000092462 UP000007801 UP000075880 UP000019118 UP000030742 UP000030765 UP000009046 UP000085678 UP000076858 UP000183832 UP000014760 UP000005408 UP000036403 UP000053097 UP000279307 UP000242188 UP000000311 UP000075809 UP000242457 UP000002358
UP000007266 UP000009192 UP000008792 UP000095300 UP000092553 UP000092443 UP000001070 UP000092444 UP000008744 UP000092445 UP000001819 UP000268350 UP000037069 UP000078200 UP000095301 UP000001292 UP000000304 UP000008711 UP000076408 UP000091820 UP000002282 UP000192221 UP000075920 UP000075883 UP000000803 UP000069272 UP000000673 UP000075885 UP000075886 UP000075881 UP000075901 UP000008820 UP000007798 UP000075884 UP000075900 UP000002320 UP000076407 UP000075903 UP000075902 UP000075840 UP000007062 UP000235965 UP000027135 UP000092462 UP000007801 UP000075880 UP000019118 UP000030742 UP000030765 UP000009046 UP000085678 UP000076858 UP000183832 UP000014760 UP000005408 UP000036403 UP000053097 UP000279307 UP000242188 UP000000311 UP000075809 UP000242457 UP000002358
Pfam
Interpro
IPR022052
Histone-bd_RBBP4_N
+ More
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR036815 14-3-3_dom_sf
IPR001848 Ribosomal_S10
IPR000308 14-3-3
IPR007632 Anoctamin
IPR019343 Unchr_KLRAQ/TTKRSYEDQ_N
IPR023409 14-3-3_CS
IPR027486 Ribosomal_S10_dom
IPR032394 Anoct_dimer
IPR036838 Ribosomal_S10_dom_sf
IPR023410 14-3-3_domain
IPR019348 KLRAQ/TTKRSYEDQ_C
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR036815 14-3-3_dom_sf
IPR001848 Ribosomal_S10
IPR000308 14-3-3
IPR007632 Anoctamin
IPR019343 Unchr_KLRAQ/TTKRSYEDQ_N
IPR023409 14-3-3_CS
IPR027486 Ribosomal_S10_dom
IPR032394 Anoct_dimer
IPR036838 Ribosomal_S10_dom_sf
IPR023410 14-3-3_domain
IPR019348 KLRAQ/TTKRSYEDQ_C
Gene 3D
ProteinModelPortal
H9JRG7
A0A212ELD8
A0A2H1WZA0
A0A3S2TSQ5
A0A2A4JKW4
A0A194R4Y3
+ More
A0A194PG85 D6WH34 B4KM61 B4LNM8 A0A1I8PI13 A0A0M4E7E4 A0A1A9YE60 A0A0M4E580 B4J5Q6 A0A1B0FG95 B4H8C7 A0A1A9ZCA6 Q28YP6 A0A3B0JXJ6 A0A0L0CBR3 A0A1A9VQJ2 A0A1I8MWQ7 T1P9E4 B4II06 B4QCP0 B3N3I8 A0A182YEL3 T1DM33 A0A1A9WWK2 A0A034VFS4 A0A1Q3EXF6 B4NYX9 A0A1L8DX10 A0A1W4WA68 A0A182WLI6 A0A0K8VTZ3 A0A182M8R8 Q8SYL1 A0A2M4BN27 A0A2M4BMA6 A0A182FMQ9 A0A2M4AEI2 W5J9U4 W8B227 A0A2M4AE82 A0A2M4AE84 A0A182P0N6 A0A2M3ZHX6 A0A182QIL7 A0A182JND6 A0A2M4BM72 A0A182SW66 Q16IG2 B4NMV5 A0A182NK56 A0A182RP63 A0A2M4BQ15 B0XBR5 A0A182WWZ4 A0A182UT22 A0A182TFD3 A0A182I2J1 Q7QIY0 Q16U37 A0A2J7RKS2 A0A067R1H4 A0A182WWZ2 A0A0K8TRP1 A0A1B0D5R1 B3MBQ2 A0A182IKF6 N6UDW6 A0A084WK73 A0A1L8DWP7 E0VGG3 A0A1Y1N8J2 A0A336MAG9 A0A1S3H902 A0A0P5ZMX4 A0A0P5S9D2 A0A1J1IKG9 A0A1J1IH46 A0A0P5VAU6 A0A0N8AP86 R7TDG9 K1QL27 A0A0J7NZ36 A0A1B6H2A8 A0A026VV92 A0A210QGC0 E2A728 A0A0P5ZAW2 A0A151X1Q0 E9IE28 A0A0P6B6T9 A0A2A3E0B9 K7J320
A0A194PG85 D6WH34 B4KM61 B4LNM8 A0A1I8PI13 A0A0M4E7E4 A0A1A9YE60 A0A0M4E580 B4J5Q6 A0A1B0FG95 B4H8C7 A0A1A9ZCA6 Q28YP6 A0A3B0JXJ6 A0A0L0CBR3 A0A1A9VQJ2 A0A1I8MWQ7 T1P9E4 B4II06 B4QCP0 B3N3I8 A0A182YEL3 T1DM33 A0A1A9WWK2 A0A034VFS4 A0A1Q3EXF6 B4NYX9 A0A1L8DX10 A0A1W4WA68 A0A182WLI6 A0A0K8VTZ3 A0A182M8R8 Q8SYL1 A0A2M4BN27 A0A2M4BMA6 A0A182FMQ9 A0A2M4AEI2 W5J9U4 W8B227 A0A2M4AE82 A0A2M4AE84 A0A182P0N6 A0A2M3ZHX6 A0A182QIL7 A0A182JND6 A0A2M4BM72 A0A182SW66 Q16IG2 B4NMV5 A0A182NK56 A0A182RP63 A0A2M4BQ15 B0XBR5 A0A182WWZ4 A0A182UT22 A0A182TFD3 A0A182I2J1 Q7QIY0 Q16U37 A0A2J7RKS2 A0A067R1H4 A0A182WWZ2 A0A0K8TRP1 A0A1B0D5R1 B3MBQ2 A0A182IKF6 N6UDW6 A0A084WK73 A0A1L8DWP7 E0VGG3 A0A1Y1N8J2 A0A336MAG9 A0A1S3H902 A0A0P5ZMX4 A0A0P5S9D2 A0A1J1IKG9 A0A1J1IH46 A0A0P5VAU6 A0A0N8AP86 R7TDG9 K1QL27 A0A0J7NZ36 A0A1B6H2A8 A0A026VV92 A0A210QGC0 E2A728 A0A0P5ZAW2 A0A151X1Q0 E9IE28 A0A0P6B6T9 A0A2A3E0B9 K7J320
PDB
3CFV
E-value=2.78386e-17,
Score=217
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Membrane
Membrane
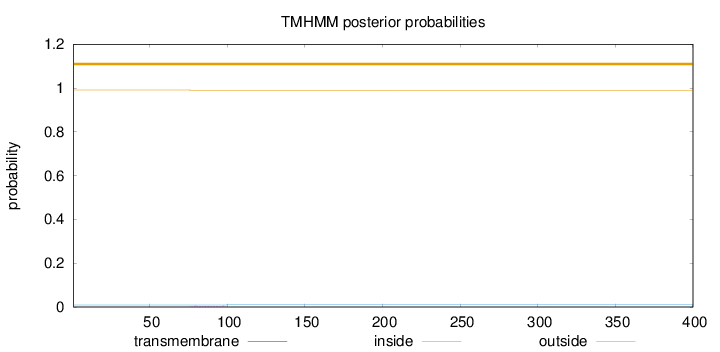
Length:
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03739
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.00901
outside
1 - 400
Population Genetic Test Statistics
Pi
252.780646
Theta
192.417689
Tajima's D
0.991966
CLR
0.000108
CSRT
0.654917254137293
Interpretation
Uncertain
