Gene
KWMTBOMO06757
Pre Gene Modal
BGIBMGA012027
Annotation
PREDICTED:_mismatch_repair_endonuclease_PMS2_isoform_X1_[Bombyx_mori]
Full name
Mismatch repair endonuclease PMS2
Alternative Name
DNA mismatch repair protein PMS2
PMS1 protein homolog 2
PMS1 protein homolog 2
Location in the cell
Nuclear Reliability : 3.958
Sequence
CDS
ATGGACAATTTAGGTACACCTTCAAGTGATATAAAACCCATAAATGAAAATACTGTTCATAAAATTTGTTCGGGACAGGTAGTTTTGAACTTGGCTGTAGCAGTTAAGGAATTAGTGGAAAATTCACTGGATGCAGGAGCTACCAAAATTGATATTAGATGCAAGAATTATGGTATGGATAGCATTGAAGTTTCAGACAATGGTTCAGGAGTTAATGAAGATAATTTTGCAGCATTGACCTTAAAATATCATACATCAAAATTGAGTGACTACTCAGATTTGCTTGGAGTATCAAGTTTTGGTTTTAGAGGTGAAGCTCTTAGTTCACTTTGTGCTTTGGCAAATCTGACTGTAACTACCAGGCATAAAAATTCCAATTATGCAATTAAAATAGAATACGATCTTAAAGGTAACATTGTAAAGACAACACCATGTTCTAGGCAAATTGGAACAACCGTTACTTGTACAAATCTGTTTCATTCCTTGCCAGTGAGACAAAAGGAATTTCATAACAATGTAAAAAGAGAATTCAACAAAATGACGCATTTACTATATGCATACTGTTTAATTTCAATAGGTGTTAAAATTACGTGTACGAATCAGGCATCTTCCAATACAAAATCTGTAGTAGTTGAAACGCATGGCTCCTCGTATAAAGACAACATTGCCAGTATATTTGGTCTTAAACAACTACAGACAATAATCGAAATAAAACCCGAATACACTCAAAACATAAAAGATAATATTTTTAAAGGTCTATCTAGTGAAGTAGCTCATGATCAACAAAGTTTTGTCATCGAAGATGTGGACATTAACCTATCAGAAGATTCCAACGACAACGAGTCTGAAGCTATTGCAAAAGATGAAATTAAATCACAAGGATATAAAAACATTGTTAAACCGTTTGAATTTATAGGTTTTGTATCTTCGTGTGAACATGGGCGTGGTAGATCTAGTACAGACAGACAGTTCTTCTTCGTAAATAACAGGCCTTGTGAACCAATTAAAATAACTAAACTCATAAATGAAGTTTACAGACAATACAATCCAAACCAATATCCTTTTGTTTTTCTGAATGTTAATGTTGAAAGATCGTCTGTAGACATAAATCTAACGCCGGATAAAAGAAAATTATTTTTAACCAAAGAGAAAAATATATTGGACGTTTTAAAAGTTTCTTTACTTAAGTTATTTGAAAGTATACCGCGGACATTGAAAGTAGACTCTTCCCAAAATGTTTCTATTTCGAAAGATCATAAACCAGAATTAGATCAACCGAGAATTTTTACGTCTTTCCTGCAACAATTCGATAAAAAATCAAAGCTTTGTGATCCTATTTCAAAAAGGGAAGATTTAAATAAAGTTGATTTAAAAAGAAAATCTACAGCAATTACTGACTACATTACTATGAAAAGTAAAAGACTAGAACAACCCTGTTCAAGTATTAATGAATCAAAATTGATGTCCAGTGATATTAATACCAATGAATTGCTTGATTCAGAAGAGATCCCTCAAGTCCCTATGAAATCTGAGGGGCGCAGTGATGACAAAAATGTTACAGAATTAATACCTCGACCAACAAATATAGAAACGCCGATAATTATATCTGACCAAATTAAATCTGAAGAAACTGAAATCACAGAGGAAAAAGAGATTATATATTTAGACTATTGCAACGAAATGCCAAATACACAGATAAAACAGTTATCAGAACAAGTTGTAGAGCAAACTTATACCATTAACTGCAAAACAAAAACAAAACCTAAGATACATTTACCACCGGGATCTAAAAAAACCTCCGAAAGCTACAATAAAGTTTTAACGGTGACAGACAAAGAAGACTTAGGTAAGAACCACAGGAGATCAGTAATTTTAAAAACTTCTTTGCAGCATGTCCAGGCTTTAACAGAAATTTATAATAGAAATAATGAAAGGATATTACCTACTAAAGTGAAATTTAAAAGCGCCATTAATCCAGTATTCAATAAAAAATGTGAAGAAGAATTGATACGAGAAATATCTAAGGATTCATTTAAGAAGATGCATATTGTGGGGCAATTTAATTTGGGTTTCATAATAACACAACTAGAAGATGATCTTTTTGTAATAGACCAACATGCTACTGATGAGATTTATAATTTTGAAACCTTACAGCGTACAACAGAACTAACCAGTCAAAAATTGGTGATCCCACAACAACTAGAGTTAACAGGTGTAAACGAAGAAATACTAATGGACAACATAGATATTTTCAAAAAGAACGGTTTTACATTTGAAGTAAATAAGGAGGCATTGCCAACGAAACGTGTAAAACTTTTGACTATACCAATGTCGAAAAATTGGTTATTTGGAAAGGAAGATATCGAAGAAATGCTCTTTATGTTAAGGGAAGCTCCGACCGAACATTGCAGGCCGAGTAGAGTAAGAGCAATGTTCGCATCGAGAGCTTGCAGAAAGTCTGTCATGATTGGCACAGCTTTGAAAAAATCCGACATGAGAACGCTGGTCGACCATATGGCAGAAATTGATAAGCCGTGGAACTGTCCACATGGACGCCCCACCATAAGGCATTTGGTCAACTTAGCTATGGTCCAAACTGAGTACAGCAGTGGATATAAATTTTGA
Protein
MDNLGTPSSDIKPINENTVHKICSGQVVLNLAVAVKELVENSLDAGATKIDIRCKNYGMDSIEVSDNGSGVNEDNFAALTLKYHTSKLSDYSDLLGVSSFGFRGEALSSLCALANLTVTTRHKNSNYAIKIEYDLKGNIVKTTPCSRQIGTTVTCTNLFHSLPVRQKEFHNNVKREFNKMTHLLYAYCLISIGVKITCTNQASSNTKSVVVETHGSSYKDNIASIFGLKQLQTIIEIKPEYTQNIKDNIFKGLSSEVAHDQQSFVIEDVDINLSEDSNDNESEAIAKDEIKSQGYKNIVKPFEFIGFVSSCEHGRGRSSTDRQFFFVNNRPCEPIKITKLINEVYRQYNPNQYPFVFLNVNVERSSVDINLTPDKRKLFLTKEKNILDVLKVSLLKLFESIPRTLKVDSSQNVSISKDHKPELDQPRIFTSFLQQFDKKSKLCDPISKREDLNKVDLKRKSTAITDYITMKSKRLEQPCSSINESKLMSSDINTNELLDSEEIPQVPMKSEGRSDDKNVTELIPRPTNIETPIIISDQIKSEETEITEEKEIIYLDYCNEMPNTQIKQLSEQVVEQTYTINCKTKTKPKIHLPPGSKKTSESYNKVLTVTDKEDLGKNHRRSVILKTSLQHVQALTEIYNRNNERILPTKVKFKSAINPVFNKKCEEELIREISKDSFKKMHIVGQFNLGFIITQLEDDLFVIDQHATDEIYNFETLQRTTELTSQKLVIPQQLELTGVNEEILMDNIDIFKKNGFTFEVNKEALPTKRVKLLTIPMSKNWLFGKEDIEEMLFMLREAPTEHCRPSRVRAMFASRACRKSVMIGTALKKSDMRTLVDHMAEIDKPWNCPHGRPTIRHLVNLAMVQTEYSSGYKF
Summary
Description
Component of the post-replicative DNA mismatch repair system (MMR). Heterodimerizes with MLH1 to form MutL alpha. DNA repair is initiated by MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of PMS2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (MLH1-PMS2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in case of major DNA damages.
Subunit
Heterodimer of PMS2 and MLH1 (MutL alpha). Forms a ternary complex with MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3). Part of the BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the RAD50-MRE11-NBS1 protein complex. This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains. Interacts with MTMR15/FAN1.
Similarity
Belongs to the DNA mismatch repair MutL/HexB family.
Keywords
Complete proteome
DNA damage
DNA repair
Endonuclease
Hydrolase
Nuclease
Nucleus
Reference proteome
Tumor suppressor
Feature
chain Mismatch repair endonuclease PMS2
Uniprot
EC Number
3.1.-.-
Pubmed
EMBL
BABH01016402
BABH01016403
NWSH01001104
PCG72591.1
ODYU01012166
SOQ58331.1
+ More
KQ460779 KPJ12299.1 RSAL01000007 RVE54098.1 JTDY01001566 KOB73512.1 GFTR01008102 JAW08324.1 DS232648 EDS44415.1 GL732526 EFX87911.1 AC121917 BC141287 CH466529 AAI41288.1 EDL19032.1 AK146365 BAE27115.1 AJWK01005056 UFQS01000700 UFQT01000700 SSX06277.1 SSX26631.1 U28724 GEDC01009645 JAS27653.1 GEDC01028846 GEDC01025856 JAS08452.1 JAS11442.1
KQ460779 KPJ12299.1 RSAL01000007 RVE54098.1 JTDY01001566 KOB73512.1 GFTR01008102 JAW08324.1 DS232648 EDS44415.1 GL732526 EFX87911.1 AC121917 BC141287 CH466529 AAI41288.1 EDL19032.1 AK146365 BAE27115.1 AJWK01005056 UFQS01000700 UFQT01000700 SSX06277.1 SSX26631.1 U28724 GEDC01009645 JAS27653.1 GEDC01028846 GEDC01025856 JAS08452.1 JAS11442.1
Proteomes
Interpro
IPR042121
MutL_C_regsub
+ More
IPR002099 DNA_mismatch_repair_N
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR014762 DNA_mismatch_repair_CS
IPR038973 MutL/Mlh/Pms
IPR013507 DNA_mismatch_S5_2-like
IPR037198 MutL_C_sf
IPR042120 MutL_C_dimsub
IPR036890 HATPase_C_sf
IPR014790 MutL_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR003594 HATPase_C
IPR002099 DNA_mismatch_repair_N
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR014762 DNA_mismatch_repair_CS
IPR038973 MutL/Mlh/Pms
IPR013507 DNA_mismatch_S5_2-like
IPR037198 MutL_C_sf
IPR042120 MutL_C_dimsub
IPR036890 HATPase_C_sf
IPR014790 MutL_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR003594 HATPase_C
ProteinModelPortal
PDB
1H7U
E-value=1.08389e-102,
Score=957
Ontologies
PATHWAY
GO
GO:0016887
GO:0030983
GO:0006298
GO:0032300
GO:0005524
GO:0004519
GO:0032389
GO:0042493
GO:0003677
GO:0032138
GO:0051321
GO:0036464
GO:0005634
GO:0005829
GO:0016446
GO:0048304
GO:0005654
GO:0015630
GO:0016447
GO:0006281
GO:0005886
GO:0006974
GO:0048298
GO:0046872
GO:0006355
GO:0043565
GO:0016567
GO:0016020
GO:0006260
PANTHER
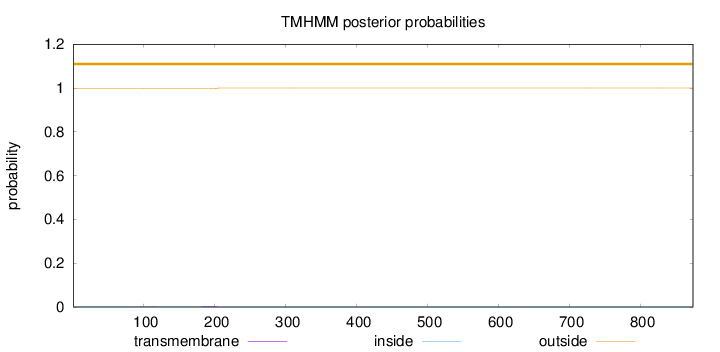
Topology
Subcellular location
Nucleus
Length:
874
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06007
Exp number, first 60 AAs:
0.00059
Total prob of N-in:
0.00218
outside
1 - 874
Population Genetic Test Statistics
Pi
242.876901
Theta
170.113645
Tajima's D
1.171039
CLR
0.141578
CSRT
0.706514674266287
Interpretation
Uncertain
