Gene
KWMTBOMO06755
Annotation
PREDICTED:_uncharacterized_protein_LOC105387844_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 2.148 Nuclear Reliability : 1.675
Sequence
CDS
ATGGTGCTCACACGGTCCGAGAAGATGAGGTCGTCACGCCTTGACCGGCCCGGCAACGAAGCTGTCTCGGGCTCAACTTTGCAAGACGCACAGCAACGAGCTGTGTCGGCCGCGCCGCGTTCGGGAGTGACGCAGGGACCGCCACCGATGGCGGCAACCTCCTCCCAGTCAACCGCGCCGCGTTCGGGAGTGACGCAGGGACCGCCACCGATGGCGGCACCCTCCTCCCAAGTGGAAAAGTGTCCGGGAGTGCCGCGGGGCTCACAAGCGATGCCCCCCTCCCGCCAAATATTGCCTACGCAATACTCGCCGGCGTCGCCAACGGGACCGAACAGCGGGGTCTGTCGTCGAGTGGTCAGACCTCCCAGCACCGCGACACCGGCAACAACTTCAGGCGACGCAGCAACGGAGAGCGTCGTATCCGCGAGCCAGAACAAGAAGGAGTCAGCGCTCGCGGCCAATCAAATAGTACACGGGTCCCACATCACGGACGCCGCTGCCAAAATCTCGACGACAGAAGAAGGCTCCACACTCCGCGACGGGATGGCGCCACCAGTACGAGGATCTTCAAGAAAGTCGAGTCAACAGTCACAACGGCTGTTGCTTATGGCGCGCCTCAAGGAAGAGCATCTTCGCGCCAAGGAAGAACAAGCCCGACTCCAAGCAGAGCTCGCCGCCGCCCGCATCTCAACATTAGAAGCCGAAGCGCTCGTTGAAGAGGAGGAAGATGCGCGCACAACACTCACGGAGGACGAGAACGAGCAATCACAGCGCATCGACACATGGCTCAGTCAGCAACGATCGATCGCGCTACACGGAGTCGAAGAAAGAGTAATGCAGCCAACACACGGATCACCAGCCACCATCGAGCAACCGCAACTGGAACTTCCCGCACCAATAGAACAGCTGAAGCTACCGGCCCCTGCAGCGGCACCGATTGCCGCACCTGTCGCACCGAAGAGCGACATCGCCGAACTAGCAGCTGCCATCGCGACGGCCGCCCGCCAAGCCAGGCCCGGACCATCGCATCGGTACTACGGGGAGCTTCCAGTGTACAGTGGATCGCATCAAGAATGGCTCTCCTTCAAGGTCTCCTACGCCGAATCAGCGGACAGCTTCAGCGCCGCAGAGAACACTGCGAGACTTCGGCGTACGTTGAGAGGAAGAGCAAGAGAAGCGGTCGAAAACTTGTTGCTGCATTACACCGAGCCCGCCGAGATCATGCGGACTCTAGAATCGCGCTTCGGACGTCCAGAAGCAATCGCCGCAACGGAGCTGGAACGACTGCGCGCGCTGCCACGCTGCACGGACACACCGAGGGACATCTGCGTATTCGCGAATAAGGTGAACAACGTCGTCGCCGCTCTGAGGGCCCTCGACCGCGTACATTACATGTACAACCCGGAGCTCACCAACATCACATCAGAGAAGCTGCCGTCCACTCTACGCCACCGCTGGTTCGAATTCTCAGCGACTCAACCGGCGGGAGAACCGGACCTCGTCAAGCTGTCGCGCTTCCTGCAACGCGAGGCGGACCTGTGCAGCCCATACGCACAGCCGGAGCCAGAGACGAGAGTGGAGAACACCAGCGGTCGGCGGAAGATGGTGAATACGCCTCAGAGGACGCACACCACGCAAGCGAAGGAAGAGAGGAAATGCGCAGCATGTCAGAAGCCGGGGCACATCCCACAAGATTGTCCCGAATTCAAGAGAGCAGCCGTCAGCGAGCGCTGGGATACGGCGAAGCGCGAAAATTTGTGTTTCCGGTGCCTACGGTTCCGGTCCCGGGGACACGTGTGTAAAAAGAAGAAGTGCGGCGTCGACAGCTGTGAACGCACGCACCACGAGCTGTTGCATAAGAAGCCAGCATGGCAGAAGCCGAAGGAAAAAGAAGAGGCGGTCACCTCGACGTGGGCCGCAAGAAGTGCGACAGCCTACCTAAAGATGGCGCCAATTACTGTTATCGGACCGGCGGGAGAAGCAGATACATGGGCCCTGCTGGACGACGGGTCAACGATCTCGCTGATTGACGAAGACTTCGCGAGACGAGTGGGCGCCAAAGGACCGATCGAACCCCTCTTCATCACTGCCATCGGAAATAACAAGATAGACGCAACACGCTCACGACGCGTCCCGCTGAAGCTATGCGGACGCACGGGGGAACCGCAAGAACTGAATCTACGGACAGTCAACGGCTTAAAATTAACACCACAGCGACTGAACATCGAAGTCCTCGCGTCGTGCCATCACCTCACCGACCTCCGCCATCACTTCAAAACGAGCTACGCCTCGCCGAAGATCCTGATCGGCCAAGACAACTGGCACCTGCTGGTGACGGAGGAGATGCGGACGGGACGACGCGATCAACCAGTGGCGTCTCGTACTCCGCTCGGGTGGGTCGTGCACGGAGCACACCCGGGAGGCAAGAGACAGCGCGTCAACTTCGTGGCACACGCGACGACGGCCGACGCGAACATGGACGAAGCCCTCAGGCACTACTTCGCGATCGAGGGACTCACCGTCGCCGCCAAGACGCCAAAGAATGACCCAGACGAGCGCGCGCTGAAGATCCTGAGAGAGACCACTCATCAGCAACCCGACGGCCGCTACGAGACCGCACTGCTGTGGCGTGAGGAGGGCCTGAAGATGCCCAACAATTTCGAAGCCGCGCTAAATCGCCTGACGTCCGTAGAGAAGAAACTGGAGAAGGATCCAAATTTGAAAGAGCGCTACAAGCAACAAATGGACGCACTGGTAGCGAAAGGGTACGCCGAAGTAGCTCCGTCAACGGGGACGAAGGACCGAACCTGGTACCTGCCGCACTTCGGCGTAACACATCCGATGAAGCCGGACAAGATTCGCATCGTGCACGATGCCGCTGCGAAGACCAGGGGGAAGAGCCTCAACGACTTCCTGCTCACCGGCCCGGACCTGCTGCAGTCTCTGCCTGGAGTGATGATGCGCTTCAGGCAGCACGCCGTCGCCGTCTCAGCGGACATCACGGAGATGTTCATGCAAATAGGCGTCCGAAGCGAGGACCGCGACGCGCTCCGGTACTTATGGAGGGAGGACCCTTCACAGGAGCCTACAGAGTACCGGATGAAATCTATAATCTTTGGCGCGACAAGCTCACCCGCGACCGCCATCTACGTGAAGAACGCCAACGCGGAGAAGTACCGACTGCAGTACCCGGACGCCGCCGACGCAATCGTACGCAACCACTATATGGACGATTACCTGGGAAGTTACCGGACAAGCGAAGACGCGATCAGAATCGCAAATGAGGTGAGGGCAGTACACCGGCAAGGCCATTTCGAGTTACGAAAATGGACCTCGAACAACGAGGAGGTACGGTGCCACATGACCGACGAAGCCCCACCTACAAGTGTGACGCTGTCCGATGGAACGGAAATAGAACGAGTACTTGGCCTGGTGTGGAGACCTGCTGAAGACGTCCTCGCCTTCAATCTGGACATGGCGCGAATCCCAAAAGAAGTCTTGGAGAGCAGCGCGCCTACGAAGAGACAGGCTCTGCGAATAATGATGTCCGTCTACGATCCCCAAGGCATCGCATCACCAGTCACTATCGGAGCGAAGAAGATCCTGCAAGAGACGTGGCGCCGAGGGACGGCGTGGGACTCCCCCCTCGATGAAGACCTCTGCGCGATGTGGAGAGAGTGGATCGACCACTTAAGACGCCTGCGCGGCGTCTCAGTGCCGCGCTGCTATCCCGGCTACTCAGACGCCGCCAACCTCCAGCTCCACACATACGTAGACGCCAGTGAGTCCGCCTACGCCGCCGCGTTATACTGGCGCGCCGAAATGCCCGACGGGGAGATCAGGACCTCCCTCGTACTCGCCAAAGCCCGAGTCGCGCCCTTGAAACTCACCTCGATACCGCGCCTCGAGCTACAAGCTGCCGTGATGGGCAGCCGCATGGCCGCGGCTGTCACAGAGGAACACGACAGAAAACCAGACGCAAGAGTGTTCTGGACAGACAGCGCCACCGTATTGACCTGGCTGCGAACGGGGTCGAGGTCGTATCGACCGTTCGTCGCTCACCGCATTGCAGCGATAGAAGAGAACAGCACTGTGGAGGAATGGCGCTGGGTCCCCACGAAGGAAAATGTCGCCGACGACGCCACTAGAGAGCTACCCGTCGGCTTTGAAAGAAACCACCGCTGGTTCATAGGCCCGGAATACCTAAGACGGCATCCGGATGATTGGCCCGTACAACGGACTGCAAGAAGGGCAGAAGAGACGGGTGAAGAGAAGTGCGCAATGCTCGCCGTGAGCAGAGAGAGCCTCGGCGAAGCGATCCCGGACCCAAGACGCTTCTCCAAGTGGGAGAAGTACCTGCGTGCAACGGGGCGAATACTACAATTCGTCAGCCTATGCCGCAGGAGTCGCGAGCGCACTCATTACAAGAGGACCAGACGGAACCCGCGTTCGGACCCGACGTGGGAGAAGAATAGAAAGAAGACGACTTCGAAGACCCCGCTGAACACACCGCGGACGCCAGAGCAAGCAGTCATTCAATGGAAGACTTTAGACGCCGACACGCTTCGACGCGCCGAGACGCTCATTTGGCGACAGAGCCAGCGGGCGGCCTTCGAGGAAGAGATTTTGACGCTCCAGCGAGGGAAACAAATATCCCCAGCGAGCCGACTGCGAAACTTGTCCGTAACCTACGCGGACGGGATATTAAGAATAAACGGACGCATCGGCAACATTAAGGGAGCTGACGTCATCACCTCGCCTCTCGTGCTAGACGGATCCCGACGCGAAACGAGACTGATGATAGACTTCATCCATAGAAAGATGCACCACGCCGGAACCGAAGCTACGATCGCCGAGTGCCGACAGTCGTACTGGGTTTTACGCCTACGCCCAGTGACACGGAGCGTAATCCACAACTGTCTTCCGTGTCGTATCCGCAAGGACACACCACCGCGTCCAGCCACGGGCGATCACCCACCGAGCAGACTGGCACACCATCGGCGACCATTTACATACGTGGGCCTCGATTACTTCGGGCCCTATCAAGTTACCACCGGCAGAAGCACCCAGAAGCACTACGTGGCCATCTTCACATGTCTCACTACGCGTGCGGTACATTTAGAACCGGCAGCGAGCCTCAGCACAGACTCAGCGGTGATGGCACTCCGGCGCATGATCGCGCGCCGGGGAGCCCCGACGGAGATATGGAGCGACAACGGCACCAATTTACGGGGTGCCGACAAGGAGCTGCGCCAAGCTTTGGACAAGGCGACTGAGCATGAAGCGAGCTTAAGAATTATCCAATGGCGCTTCATCCCACCGGGCGCGCCTTTCATGGGCGGCGCCTGGGAAAGAATGGTACGGGCAGTAAAGGCCGCGCTCTCGGCCACGGAGCAGCCGAGGCACCCGACGCCCGAAATATTTCACACTCTGCTAGCGGAGGCAGAGTTCACAGTGAACAGCCGACCTCTCACCCACGTATCGGTGAGCGCCGACGATCCCGACCCTCTGACACCGAACCACTTCCTGCTGGGAGGCCCGGCGCGAGTCCCCGTGCCGGGCAAGTTTGACGACGCCGACCTCATAGGGCGCGCACACTGGCGCGCAGCTCAACGTCTGGCAGATGTGTTCTGGAGTCGCTGGCTCCGGGAGTACCTGCCCGACCTGCAGAACCGGCGGGAGCCCCACAGCCGCGGGCCCGCCCTGAAGATAGGCGACGTCGTGATAATAGCAGACGGAACGCTCCCACGGAACACCTGGCCACGTGGGATCATCCAGGAGGTTTACCCGGGGGCCGACGGCATCACTCGAGTGGTCGACGTGCGCACCGCCGGCGGTATATTGCGACGCCCGGCAAAGAAGATTATCGTGCTGCCCACGATGACCGCCGCTCACCAGGGAGGATGCAGCGACGTGAACGACGCTGCACGGCGGGAGGATGTTCGCGACGGACATGAATAA
Protein
MVLTRSEKMRSSRLDRPGNEAVSGSTLQDAQQRAVSAAPRSGVTQGPPPMAATSSQSTAPRSGVTQGPPPMAAPSSQVEKCPGVPRGSQAMPPSRQILPTQYSPASPTGPNSGVCRRVVRPPSTATPATTSGDAATESVVSASQNKKESALAANQIVHGSHITDAAAKISTTEEGSTLRDGMAPPVRGSSRKSSQQSQRLLLMARLKEEHLRAKEEQARLQAELAAARISTLEAEALVEEEEDARTTLTEDENEQSQRIDTWLSQQRSIALHGVEERVMQPTHGSPATIEQPQLELPAPIEQLKLPAPAAAPIAAPVAPKSDIAELAAAIATAARQARPGPSHRYYGELPVYSGSHQEWLSFKVSYAESADSFSAAENTARLRRTLRGRAREAVENLLLHYTEPAEIMRTLESRFGRPEAIAATELERLRALPRCTDTPRDICVFANKVNNVVAALRALDRVHYMYNPELTNITSEKLPSTLRHRWFEFSATQPAGEPDLVKLSRFLQREADLCSPYAQPEPETRVENTSGRRKMVNTPQRTHTTQAKEERKCAACQKPGHIPQDCPEFKRAAVSERWDTAKRENLCFRCLRFRSRGHVCKKKKCGVDSCERTHHELLHKKPAWQKPKEKEEAVTSTWAARSATAYLKMAPITVIGPAGEADTWALLDDGSTISLIDEDFARRVGAKGPIEPLFITAIGNNKIDATRSRRVPLKLCGRTGEPQELNLRTVNGLKLTPQRLNIEVLASCHHLTDLRHHFKTSYASPKILIGQDNWHLLVTEEMRTGRRDQPVASRTPLGWVVHGAHPGGKRQRVNFVAHATTADANMDEALRHYFAIEGLTVAAKTPKNDPDERALKILRETTHQQPDGRYETALLWREEGLKMPNNFEAALNRLTSVEKKLEKDPNLKERYKQQMDALVAKGYAEVAPSTGTKDRTWYLPHFGVTHPMKPDKIRIVHDAAAKTRGKSLNDFLLTGPDLLQSLPGVMMRFRQHAVAVSADITEMFMQIGVRSEDRDALRYLWREDPSQEPTEYRMKSIIFGATSSPATAIYVKNANAEKYRLQYPDAADAIVRNHYMDDYLGSYRTSEDAIRIANEVRAVHRQGHFELRKWTSNNEEVRCHMTDEAPPTSVTLSDGTEIERVLGLVWRPAEDVLAFNLDMARIPKEVLESSAPTKRQALRIMMSVYDPQGIASPVTIGAKKILQETWRRGTAWDSPLDEDLCAMWREWIDHLRRLRGVSVPRCYPGYSDAANLQLHTYVDASESAYAAALYWRAEMPDGEIRTSLVLAKARVAPLKLTSIPRLELQAAVMGSRMAAAVTEEHDRKPDARVFWTDSATVLTWLRTGSRSYRPFVAHRIAAIEENSTVEEWRWVPTKENVADDATRELPVGFERNHRWFIGPEYLRRHPDDWPVQRTARRAEETGEEKCAMLAVSRESLGEAIPDPRRFSKWEKYLRATGRILQFVSLCRRSRERTHYKRTRRNPRSDPTWEKNRKKTTSKTPLNTPRTPEQAVIQWKTLDADTLRRAETLIWRQSQRAAFEEEILTLQRGKQISPASRLRNLSVTYADGILRINGRIGNIKGADVITSPLVLDGSRRETRLMIDFIHRKMHHAGTEATIAECRQSYWVLRLRPVTRSVIHNCLPCRIRKDTPPRPATGDHPPSRLAHHRRPFTYVGLDYFGPYQVTTGRSTQKHYVAIFTCLTTRAVHLEPAASLSTDSAVMALRRMIARRGAPTEIWSDNGTNLRGADKELRQALDKATEHEASLRIIQWRFIPPGAPFMGGAWERMVRAVKAALSATEQPRHPTPEIFHTLLAEAEFTVNSRPLTHVSVSADDPDPLTPNHFLLGGPARVPVPGKFDDADLIGRAHWRAAQRLADVFWSRWLREYLPDLQNRREPHSRGPALKIGDVVIIADGTLPRNTWPRGIIQEVYPGADGITRVVDVRTAGGILRRPAKKIIVLPTMTAAHQGGCSDVNDAARREDVRDGHE
Summary
Uniprot
Q2MGA5
A0A3S2N5J7
A0A0N1IGH3
A0A226E2P7
A0A226EEI4
A0A226DH22
+ More
A0A226DM06 A0A226EKF8 A0A182G158 A0A182HCB4 A0A1Y1LN76 A0A226D351 A0A146KYV8 W8AJC8 A0A0K3CPX7 A0A182H5B8 A0A182HDR0 A0A2M4CKI9 A0A1W7R681 A0A226EDU9 A0A2M4CKZ0 A0A182G1E0 A0A226EFR1 A0A2M4B8T6 A0A2M4B8V2 A0A1W7R6F2 A0A1W7R6A0 A0A1W7R6D5 A0A1W7R6F7 Q76C94 Q76C95 A0A2M4CVY8 A0A2M4B8I4 A0A085NQS3 A0A2M4B8T5 W4Z131 W4Y2R6 W4XIU6 W4ZGF8 W4XIU3 W4XIU5 A0A0N5E779 W4XEJ9 W4XIU4 W4ZKP7 W4ZGF6 W4ZGF3 W4XEJ7 A0A0P6AZF3 A0A0P6C5E3 W4XEJ5 A0A0V1MV06 A0A0V1GTE8 A0A0N5E0H7 A0A2M4B963 A0A0V1MUH2 A0A2M4B9W9 A0A2M4B9D0 A0A2M4B9E1 A0A1S3I2B0 A0A0P6AFC1 A0A2H9T496 A0A0P6IXJ2 A0A2B4R8F3 A0A0P6IQY0 A0A0V0WCL3 A0A0N5E6S2 A0A0V0RN93 A0A0V0RR42 A0A0V1C9I9 A0A0V1F871 A0A0V1GWL8 A0A0N5DLM5 A0A0V0T6P1 W4ZGF9 A0A146TD72 A0A0V1AZZ2 A0A085N448 A0A0V0ULI0 A0A0V1G1T1 W4ZF56 A0A0V0XF38 A0A0V1GX78 W4ZF57 A0A2B4RJC8 A0A0V1NDE4
A0A226DM06 A0A226EKF8 A0A182G158 A0A182HCB4 A0A1Y1LN76 A0A226D351 A0A146KYV8 W8AJC8 A0A0K3CPX7 A0A182H5B8 A0A182HDR0 A0A2M4CKI9 A0A1W7R681 A0A226EDU9 A0A2M4CKZ0 A0A182G1E0 A0A226EFR1 A0A2M4B8T6 A0A2M4B8V2 A0A1W7R6F2 A0A1W7R6A0 A0A1W7R6D5 A0A1W7R6F7 Q76C94 Q76C95 A0A2M4CVY8 A0A2M4B8I4 A0A085NQS3 A0A2M4B8T5 W4Z131 W4Y2R6 W4XIU6 W4ZGF8 W4XIU3 W4XIU5 A0A0N5E779 W4XEJ9 W4XIU4 W4ZKP7 W4ZGF6 W4ZGF3 W4XEJ7 A0A0P6AZF3 A0A0P6C5E3 W4XEJ5 A0A0V1MV06 A0A0V1GTE8 A0A0N5E0H7 A0A2M4B963 A0A0V1MUH2 A0A2M4B9W9 A0A2M4B9D0 A0A2M4B9E1 A0A1S3I2B0 A0A0P6AFC1 A0A2H9T496 A0A0P6IXJ2 A0A2B4R8F3 A0A0P6IQY0 A0A0V0WCL3 A0A0N5E6S2 A0A0V0RN93 A0A0V0RR42 A0A0V1C9I9 A0A0V1F871 A0A0V1GWL8 A0A0N5DLM5 A0A0V0T6P1 W4ZGF9 A0A146TD72 A0A0V1AZZ2 A0A085N448 A0A0V0ULI0 A0A0V1G1T1 W4ZF56 A0A0V0XF38 A0A0V1GX78 W4ZF57 A0A2B4RJC8 A0A0V1NDE4
EMBL
AF530470
AAQ09229.1
RSAL01000375
RVE42106.1
KQ459986
KPJ18861.1
+ More
LNIX01000007 OXA52015.1 LNIX01000004 OXA55241.1 LNIX01000019 OXA44855.1 LNIX01000015 OXA46575.1 LNIX01000003 OXA57597.1 JXUM01136032 KQ568367 KXJ69026.1 JXUM01127084 KQ567147 KXJ69602.1 GEZM01052586 JAV74401.1 LNIX01000039 OXA39294.1 GDHC01018323 JAQ00306.1 GAMC01017930 JAB88625.1 LN877231 CTR11689.1 JXUM01111221 KQ565482 KXJ70922.1 JXUM01130316 KQ567562 KXJ69370.1 GGFL01001682 MBW65860.1 GEHC01000967 JAV46678.1 OXA55408.1 GGFL01001842 MBW66020.1 JXUM01038372 KQ561162 KXJ79503.1 OXA56382.1 GGFJ01000306 MBW49447.1 GGFJ01000333 MBW49474.1 GEHC01000913 JAV46732.1 GEHC01000947 JAV46698.1 GEHC01000921 JAV46724.1 GEHC01000945 JAV46700.1 AB110069 BAD01590.1 BAD01589.1 GGFL01005193 MBW69371.1 GGFJ01000218 MBW49359.1 KL367480 KFD71819.1 GGFJ01000302 MBW49443.1 AAGJ04040339 AAGJ04043389 AAGJ04001285 AAGJ04058558 AAGJ04115911 AAGJ04091218 AAGJ04095911 GDIP01036233 JAM67482.1 GDIP01004568 JAM99147.1 JYDO01000040 KRZ75294.1 JYDP01000296 KRZ01365.1 GGFJ01000432 MBW49573.1 KRZ75306.1 GGFJ01000487 MBW49628.1 GGFJ01000471 MBW49612.1 GGFJ01000470 MBW49611.1 GDIP01030392 JAM73323.1 NSIT01000285 PJE78034.1 GDIQ01002920 JAN91817.1 LSMT01001301 PFX12555.1 GDIQ01002921 JAN91816.1 JYDK01000180 KRX73317.1 JYDL01000119 KRX15895.1 JYDL01000102 KRX16693.1 JYDI01000333 KRY45736.1 JYDT01000182 KRY82272.1 JYDP01000218 KRZ02721.1 JYDJ01000536 KRX34640.1 GCES01095441 JAQ90881.1 JYDH01000144 KRY30334.1 KL367558 KFD64244.1 JYDN01000258 KRX52286.1 JYDT01000008 KRY92224.1 AAGJ04094839 JYDU01000375 KRX86446.1 JYDS01000555 KRZ02797.1 LSMT01000554 PFX16342.1 JYDM01000410 KRZ82006.1
LNIX01000007 OXA52015.1 LNIX01000004 OXA55241.1 LNIX01000019 OXA44855.1 LNIX01000015 OXA46575.1 LNIX01000003 OXA57597.1 JXUM01136032 KQ568367 KXJ69026.1 JXUM01127084 KQ567147 KXJ69602.1 GEZM01052586 JAV74401.1 LNIX01000039 OXA39294.1 GDHC01018323 JAQ00306.1 GAMC01017930 JAB88625.1 LN877231 CTR11689.1 JXUM01111221 KQ565482 KXJ70922.1 JXUM01130316 KQ567562 KXJ69370.1 GGFL01001682 MBW65860.1 GEHC01000967 JAV46678.1 OXA55408.1 GGFL01001842 MBW66020.1 JXUM01038372 KQ561162 KXJ79503.1 OXA56382.1 GGFJ01000306 MBW49447.1 GGFJ01000333 MBW49474.1 GEHC01000913 JAV46732.1 GEHC01000947 JAV46698.1 GEHC01000921 JAV46724.1 GEHC01000945 JAV46700.1 AB110069 BAD01590.1 BAD01589.1 GGFL01005193 MBW69371.1 GGFJ01000218 MBW49359.1 KL367480 KFD71819.1 GGFJ01000302 MBW49443.1 AAGJ04040339 AAGJ04043389 AAGJ04001285 AAGJ04058558 AAGJ04115911 AAGJ04091218 AAGJ04095911 GDIP01036233 JAM67482.1 GDIP01004568 JAM99147.1 JYDO01000040 KRZ75294.1 JYDP01000296 KRZ01365.1 GGFJ01000432 MBW49573.1 KRZ75306.1 GGFJ01000487 MBW49628.1 GGFJ01000471 MBW49612.1 GGFJ01000470 MBW49611.1 GDIP01030392 JAM73323.1 NSIT01000285 PJE78034.1 GDIQ01002920 JAN91817.1 LSMT01001301 PFX12555.1 GDIQ01002921 JAN91816.1 JYDK01000180 KRX73317.1 JYDL01000119 KRX15895.1 JYDL01000102 KRX16693.1 JYDI01000333 KRY45736.1 JYDT01000182 KRY82272.1 JYDP01000218 KRZ02721.1 JYDJ01000536 KRX34640.1 GCES01095441 JAQ90881.1 JYDH01000144 KRY30334.1 KL367558 KFD64244.1 JYDN01000258 KRX52286.1 JYDT01000008 KRY92224.1 AAGJ04094839 JYDU01000375 KRX86446.1 JYDS01000555 KRZ02797.1 LSMT01000554 PFX16342.1 JYDM01000410 KRZ82006.1
Proteomes
Pfam
PF03564 DUF1759
+ More
PF05380 Peptidase_A17
PF00665 rve
PF18701 DUF5641
PF05585 DUF1758
PF17921 Integrase_H2C2
PF01565 FAD_binding_4
PF09129 Chol_subst-bind
PF00628 PHD
PF00078 RVT_1
PF01048 PNP_UDP_1
PF13855 LRR_8
PF18738 HEPN_DZIP3
PF14943 MRP-S26
PF00271 Helicase_C
PF00270 DEAD
PF17919 RT_RNaseH_2
PF05380 Peptidase_A17
PF00665 rve
PF18701 DUF5641
PF05585 DUF1758
PF17921 Integrase_H2C2
PF01565 FAD_binding_4
PF09129 Chol_subst-bind
PF00628 PHD
PF00078 RVT_1
PF01048 PNP_UDP_1
PF13855 LRR_8
PF18738 HEPN_DZIP3
PF14943 MRP-S26
PF00271 Helicase_C
PF00270 DEAD
PF17919 RT_RNaseH_2
Interpro
IPR040676
DUF5641
+ More
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR002999 Tudor
IPR001841 Znf_RING
IPR000477 RT_dom
IPR000253 FHA_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR003961 FN3_dom
IPR035994 Nucleoside_phosphorylase_sf
IPR000845 Nucleoside_phosphorylase_d
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR041249 HEPN_DZIP3
IPR003591 Leu-rich_rpt_typical-subtyp
IPR027417 P-loop_NTPase
IPR026140 MRPS26
IPR013026 TPR-contain_dom
IPR011545 DEAD/DEAH_box_helicase_dom
IPR019734 TPR_repeat
IPR001650 Helicase_C
IPR011990 TPR-like_helical_dom_sf
IPR014001 Helicase_ATP-bd
IPR041577 RT_RNaseH_2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR019786 Zinc_finger_PHD-type_CS
IPR002999 Tudor
IPR001841 Znf_RING
IPR000477 RT_dom
IPR000253 FHA_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR003961 FN3_dom
IPR035994 Nucleoside_phosphorylase_sf
IPR000845 Nucleoside_phosphorylase_d
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR041249 HEPN_DZIP3
IPR003591 Leu-rich_rpt_typical-subtyp
IPR027417 P-loop_NTPase
IPR026140 MRPS26
IPR013026 TPR-contain_dom
IPR011545 DEAD/DEAH_box_helicase_dom
IPR019734 TPR_repeat
IPR001650 Helicase_C
IPR011990 TPR-like_helical_dom_sf
IPR014001 Helicase_ATP-bd
IPR041577 RT_RNaseH_2
SUPFAM
Gene 3D
ProteinModelPortal
Q2MGA5
A0A3S2N5J7
A0A0N1IGH3
A0A226E2P7
A0A226EEI4
A0A226DH22
+ More
A0A226DM06 A0A226EKF8 A0A182G158 A0A182HCB4 A0A1Y1LN76 A0A226D351 A0A146KYV8 W8AJC8 A0A0K3CPX7 A0A182H5B8 A0A182HDR0 A0A2M4CKI9 A0A1W7R681 A0A226EDU9 A0A2M4CKZ0 A0A182G1E0 A0A226EFR1 A0A2M4B8T6 A0A2M4B8V2 A0A1W7R6F2 A0A1W7R6A0 A0A1W7R6D5 A0A1W7R6F7 Q76C94 Q76C95 A0A2M4CVY8 A0A2M4B8I4 A0A085NQS3 A0A2M4B8T5 W4Z131 W4Y2R6 W4XIU6 W4ZGF8 W4XIU3 W4XIU5 A0A0N5E779 W4XEJ9 W4XIU4 W4ZKP7 W4ZGF6 W4ZGF3 W4XEJ7 A0A0P6AZF3 A0A0P6C5E3 W4XEJ5 A0A0V1MV06 A0A0V1GTE8 A0A0N5E0H7 A0A2M4B963 A0A0V1MUH2 A0A2M4B9W9 A0A2M4B9D0 A0A2M4B9E1 A0A1S3I2B0 A0A0P6AFC1 A0A2H9T496 A0A0P6IXJ2 A0A2B4R8F3 A0A0P6IQY0 A0A0V0WCL3 A0A0N5E6S2 A0A0V0RN93 A0A0V0RR42 A0A0V1C9I9 A0A0V1F871 A0A0V1GWL8 A0A0N5DLM5 A0A0V0T6P1 W4ZGF9 A0A146TD72 A0A0V1AZZ2 A0A085N448 A0A0V0ULI0 A0A0V1G1T1 W4ZF56 A0A0V0XF38 A0A0V1GX78 W4ZF57 A0A2B4RJC8 A0A0V1NDE4
A0A226DM06 A0A226EKF8 A0A182G158 A0A182HCB4 A0A1Y1LN76 A0A226D351 A0A146KYV8 W8AJC8 A0A0K3CPX7 A0A182H5B8 A0A182HDR0 A0A2M4CKI9 A0A1W7R681 A0A226EDU9 A0A2M4CKZ0 A0A182G1E0 A0A226EFR1 A0A2M4B8T6 A0A2M4B8V2 A0A1W7R6F2 A0A1W7R6A0 A0A1W7R6D5 A0A1W7R6F7 Q76C94 Q76C95 A0A2M4CVY8 A0A2M4B8I4 A0A085NQS3 A0A2M4B8T5 W4Z131 W4Y2R6 W4XIU6 W4ZGF8 W4XIU3 W4XIU5 A0A0N5E779 W4XEJ9 W4XIU4 W4ZKP7 W4ZGF6 W4ZGF3 W4XEJ7 A0A0P6AZF3 A0A0P6C5E3 W4XEJ5 A0A0V1MV06 A0A0V1GTE8 A0A0N5E0H7 A0A2M4B963 A0A0V1MUH2 A0A2M4B9W9 A0A2M4B9D0 A0A2M4B9E1 A0A1S3I2B0 A0A0P6AFC1 A0A2H9T496 A0A0P6IXJ2 A0A2B4R8F3 A0A0P6IQY0 A0A0V0WCL3 A0A0N5E6S2 A0A0V0RN93 A0A0V0RR42 A0A0V1C9I9 A0A0V1F871 A0A0V1GWL8 A0A0N5DLM5 A0A0V0T6P1 W4ZGF9 A0A146TD72 A0A0V1AZZ2 A0A085N448 A0A0V0ULI0 A0A0V1G1T1 W4ZF56 A0A0V0XF38 A0A0V1GX78 W4ZF57 A0A2B4RJC8 A0A0V1NDE4
Ontologies
GO
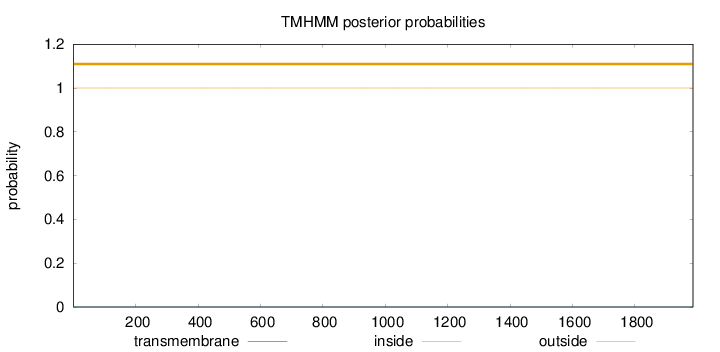
Topology
Length:
1986
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00123
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1986
Population Genetic Test Statistics
Pi
33.241527
Theta
42.076695
Tajima's D
-0.676674
CLR
0.612647
CSRT
0.206139693015349
Interpretation
Uncertain
