Gene
KWMTBOMO06748
Pre Gene Modal
BGIBMGA012024
Annotation
PREDICTED:_methyltransferase-like_protein_9_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.781
Sequence
CDS
ATGTTGAAACAAGCTCGAGCCGCGATCGCTCCTGGGGGCGTTTTACTCGTGGCGCTCGTCTTACCTTACAAGCCTTACGTTGAGGCGACCTCAGACCATAAGCCTGAGGAGCGTCTTCCGATCTACGACTGCTGCGAGTTCGAAGACCAGGTGACGGCCTTCGTGTCGTTCATGGAGAAGGAAGCCGGCTACGAGCTGGTCTCCTGGACGCGGACGCCGTACCTCTGCGAGGGAGACTTCGCTCAGGCCTACTACTGGCTCGACGACTCCGTATACGTGTTCAAGCCTAAAGCCATCCCATCCGAATGA
Protein
MLKQARAAIAPGGVLLVALVLPYKPYVEATSDHKPEERLPIYDCCEFEDQVTAFVSFMEKEAGYELVSWTRTPYLCEGDFAQAYYWLDDSVYVFKPKAIPSE
Summary
Uniprot
H9JR64
A0A2H1WW46
A0A2A4JRY1
A0A194PEC7
A0A194R8U0
A0A212F729
+ More
A0A3S2P0T4 A0A0L7LVI7 D6WH75 A0A1Y1KKD3 J3JYB1 A0A0V0GB22 A0A224XPH0 T1ICR0 A0A023F201 A0A1W4WKL0 A0A146M735 A0A2J7PDD1 A0A2P8YN48 A0A0A9YH30 A0A0K8SEQ3 J9JJX0 A0A067RBQ3 A0A068WGV8 W6UH69 A0A068Y522 A0A1D2NEB4 A0A1S3HYC4 A0A074Z2X9 A0A2R7X1R6 A0A183ALL3 A0A2G8JQ22 A0A1B6M475 A0A1S8WV56 A0A1B6GEN1 T1J491 A0A1B6HNT8 A0A210QH13 A0A2S2NII4 Q5DI52 G7YR01 A0A0X3PYL8 A0A183RSA0 A0A0P4WDQ5 A0A3P6TSE9 A0A154PN31 A0A0X3PSH1 A0A3R7DGC2 A0A2H8TID1 A0A0R3TNC8 A0A183MYI8 A0A164LXC1 A0A310SLE7 G4VPT4 E2C4W7 A0A1Q3G289 A0A183P0Z4 A0A095CC93 A0A0C9QE78 A0A2A3E547 V9KWY0 A0A0P5DQL5 A0A0P5B8X8 A0A3R7Q020 A0A0P5BTE6 A0A0N8A2W8 A0A0N8DFG3 F4W5Y6 A0A0G4GQI5 A0A0P6HC18 A0A0P6BX82 A0A0P5VHB0 A0A026WHA9 A0A0J7L2A7 B4GCY3 E2B1R2 V9L595 A0A3L8DDS0 A0A0P5AY58 A0A183SQC8 V4B3E1 Q15ER9 H3B4S5 A0A151XH67 A0A0P6GEQ7 A0A195DFK1 A0A088AQV6
A0A3S2P0T4 A0A0L7LVI7 D6WH75 A0A1Y1KKD3 J3JYB1 A0A0V0GB22 A0A224XPH0 T1ICR0 A0A023F201 A0A1W4WKL0 A0A146M735 A0A2J7PDD1 A0A2P8YN48 A0A0A9YH30 A0A0K8SEQ3 J9JJX0 A0A067RBQ3 A0A068WGV8 W6UH69 A0A068Y522 A0A1D2NEB4 A0A1S3HYC4 A0A074Z2X9 A0A2R7X1R6 A0A183ALL3 A0A2G8JQ22 A0A1B6M475 A0A1S8WV56 A0A1B6GEN1 T1J491 A0A1B6HNT8 A0A210QH13 A0A2S2NII4 Q5DI52 G7YR01 A0A0X3PYL8 A0A183RSA0 A0A0P4WDQ5 A0A3P6TSE9 A0A154PN31 A0A0X3PSH1 A0A3R7DGC2 A0A2H8TID1 A0A0R3TNC8 A0A183MYI8 A0A164LXC1 A0A310SLE7 G4VPT4 E2C4W7 A0A1Q3G289 A0A183P0Z4 A0A095CC93 A0A0C9QE78 A0A2A3E547 V9KWY0 A0A0P5DQL5 A0A0P5B8X8 A0A3R7Q020 A0A0P5BTE6 A0A0N8A2W8 A0A0N8DFG3 F4W5Y6 A0A0G4GQI5 A0A0P6HC18 A0A0P6BX82 A0A0P5VHB0 A0A026WHA9 A0A0J7L2A7 B4GCY3 E2B1R2 V9L595 A0A3L8DDS0 A0A0P5AY58 A0A183SQC8 V4B3E1 Q15ER9 H3B4S5 A0A151XH67 A0A0P6GEQ7 A0A195DFK1 A0A088AQV6
Pubmed
EMBL
BABH01016416
ODYU01011506
SOQ57281.1
NWSH01000797
PCG74162.1
KQ459606
+ More
KPI91706.1 KQ460779 KPJ12291.1 AGBW02009946 OWR49528.1 RSAL01000007 RVE54091.1 JTDY01000007 KOB79492.1 KQ971330 EFA00981.1 GEZM01081127 JAV61862.1 BT128236 AEE63196.1 GECL01000812 JAP05312.1 GFTR01004698 JAW11728.1 ACPB03013477 GBBI01003626 JAC15086.1 GDHC01003380 JAQ15249.1 NEVH01026391 PNF14332.1 PYGN01000476 PSN45671.1 GBHO01012674 GBHO01012673 JAG30930.1 JAG30931.1 GBRD01014058 JAG51768.1 ABLF02041202 ABLF02041232 ABLF02041233 KK852609 KDR20299.1 LK028579 CDS19315.1 APAU02000031 EUB60386.1 LN902843 CDS37367.1 LJIJ01000078 ODN03316.1 KL597122 KER19882.1 KK856091 PTY24835.1 UZAN01045166 VDP82182.1 MRZV01001436 PIK37881.1 GEBQ01009257 JAT30720.1 KV894317 OON18308.1 GECZ01008898 JAS60871.1 JH431841 GECU01031368 JAS76338.1 NEDP02003738 OWF48006.1 GGMR01004371 MBY16990.1 AY812772 AAW24504.1 DF144005 GAA55381.1 GEEE01006955 JAP56270.1 GDRN01045652 JAI66833.1 UYRU01045001 VDK88347.1 KQ434984 KZC13137.1 GEEE01010258 JAP52967.1 NIRI01000998 RJW68367.1 GFXV01002098 MBW13903.1 UZAE01012414 VDO05048.1 UZAI01018578 VDP38340.1 LRGB01003123 KZS04524.1 KQ762903 OAD55265.1 HE601631 CCD82440.1 GL452660 EFN77025.1 GFDL01001125 JAV33920.1 UZAL01028639 VDP42603.1 KL251389 KGB40173.1 GBYB01010030 GBYB01012708 JAG79797.1 JAG82475.1 KZ288387 PBC26412.1 JW870353 AFP02871.1 GDIP01169578 JAJ53824.1 GDIP01187792 JAJ35610.1 QCYY01003151 ROT65039.1 GDIP01180885 JAJ42517.1 GDIP01187793 JAJ35609.1 GDIP01038943 JAM64772.1 GL887695 EGI70462.1 CDMZ01001443 CEM32711.1 GDIQ01021346 JAN73391.1 GDIP01008853 JAM94862.1 GDIP01100194 JAM03521.1 KK107231 EZA55051.1 LBMM01001293 KMQ96569.1 CH479181 EDW31521.1 GL444953 EFN60418.1 JW874264 AFP06781.1 QOIP01000009 RLU18471.1 GDIP01192792 JAJ30610.1 UYSU01033684 VDL92811.1 KB203854 ESO82859.1 DQ480557 ABG21832.1 AFYH01092628 AFYH01092629 AFYH01092630 AFYH01092631 KQ982130 KYQ59743.1 GDIQ01054128 JAN40609.1 KQ980903 KYN11670.1
KPI91706.1 KQ460779 KPJ12291.1 AGBW02009946 OWR49528.1 RSAL01000007 RVE54091.1 JTDY01000007 KOB79492.1 KQ971330 EFA00981.1 GEZM01081127 JAV61862.1 BT128236 AEE63196.1 GECL01000812 JAP05312.1 GFTR01004698 JAW11728.1 ACPB03013477 GBBI01003626 JAC15086.1 GDHC01003380 JAQ15249.1 NEVH01026391 PNF14332.1 PYGN01000476 PSN45671.1 GBHO01012674 GBHO01012673 JAG30930.1 JAG30931.1 GBRD01014058 JAG51768.1 ABLF02041202 ABLF02041232 ABLF02041233 KK852609 KDR20299.1 LK028579 CDS19315.1 APAU02000031 EUB60386.1 LN902843 CDS37367.1 LJIJ01000078 ODN03316.1 KL597122 KER19882.1 KK856091 PTY24835.1 UZAN01045166 VDP82182.1 MRZV01001436 PIK37881.1 GEBQ01009257 JAT30720.1 KV894317 OON18308.1 GECZ01008898 JAS60871.1 JH431841 GECU01031368 JAS76338.1 NEDP02003738 OWF48006.1 GGMR01004371 MBY16990.1 AY812772 AAW24504.1 DF144005 GAA55381.1 GEEE01006955 JAP56270.1 GDRN01045652 JAI66833.1 UYRU01045001 VDK88347.1 KQ434984 KZC13137.1 GEEE01010258 JAP52967.1 NIRI01000998 RJW68367.1 GFXV01002098 MBW13903.1 UZAE01012414 VDO05048.1 UZAI01018578 VDP38340.1 LRGB01003123 KZS04524.1 KQ762903 OAD55265.1 HE601631 CCD82440.1 GL452660 EFN77025.1 GFDL01001125 JAV33920.1 UZAL01028639 VDP42603.1 KL251389 KGB40173.1 GBYB01010030 GBYB01012708 JAG79797.1 JAG82475.1 KZ288387 PBC26412.1 JW870353 AFP02871.1 GDIP01169578 JAJ53824.1 GDIP01187792 JAJ35610.1 QCYY01003151 ROT65039.1 GDIP01180885 JAJ42517.1 GDIP01187793 JAJ35609.1 GDIP01038943 JAM64772.1 GL887695 EGI70462.1 CDMZ01001443 CEM32711.1 GDIQ01021346 JAN73391.1 GDIP01008853 JAM94862.1 GDIP01100194 JAM03521.1 KK107231 EZA55051.1 LBMM01001293 KMQ96569.1 CH479181 EDW31521.1 GL444953 EFN60418.1 JW874264 AFP06781.1 QOIP01000009 RLU18471.1 GDIP01192792 JAJ30610.1 UYSU01033684 VDL92811.1 KB203854 ESO82859.1 DQ480557 ABG21832.1 AFYH01092628 AFYH01092629 AFYH01092630 AFYH01092631 KQ982130 KYQ59743.1 GDIQ01054128 JAN40609.1 KQ980903 KYN11670.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000007266 UP000015103 UP000192223 UP000235965 UP000245037 UP000007819 UP000027135 UP000019149 UP000017246 UP000094527 UP000085678 UP000050740 UP000230750 UP000242188 UP000050792 UP000076502 UP000046398 UP000278807 UP000050790 UP000277204 UP000076858 UP000008854 UP000008237 UP000050791 UP000242457 UP000283509 UP000007755 UP000053097 UP000036403 UP000008744 UP000000311 UP000279307 UP000050788 UP000275846 UP000030746 UP000008672 UP000075809 UP000078492 UP000005203
UP000037510 UP000007266 UP000015103 UP000192223 UP000235965 UP000245037 UP000007819 UP000027135 UP000019149 UP000017246 UP000094527 UP000085678 UP000050740 UP000230750 UP000242188 UP000050792 UP000076502 UP000046398 UP000278807 UP000050790 UP000277204 UP000076858 UP000008854 UP000008237 UP000050791 UP000242457 UP000283509 UP000007755 UP000053097 UP000036403 UP000008744 UP000000311 UP000279307 UP000050788 UP000275846 UP000030746 UP000008672 UP000075809 UP000078492 UP000005203
PRIDE
Pfam
PF05219 DREV
Interpro
ProteinModelPortal
H9JR64
A0A2H1WW46
A0A2A4JRY1
A0A194PEC7
A0A194R8U0
A0A212F729
+ More
A0A3S2P0T4 A0A0L7LVI7 D6WH75 A0A1Y1KKD3 J3JYB1 A0A0V0GB22 A0A224XPH0 T1ICR0 A0A023F201 A0A1W4WKL0 A0A146M735 A0A2J7PDD1 A0A2P8YN48 A0A0A9YH30 A0A0K8SEQ3 J9JJX0 A0A067RBQ3 A0A068WGV8 W6UH69 A0A068Y522 A0A1D2NEB4 A0A1S3HYC4 A0A074Z2X9 A0A2R7X1R6 A0A183ALL3 A0A2G8JQ22 A0A1B6M475 A0A1S8WV56 A0A1B6GEN1 T1J491 A0A1B6HNT8 A0A210QH13 A0A2S2NII4 Q5DI52 G7YR01 A0A0X3PYL8 A0A183RSA0 A0A0P4WDQ5 A0A3P6TSE9 A0A154PN31 A0A0X3PSH1 A0A3R7DGC2 A0A2H8TID1 A0A0R3TNC8 A0A183MYI8 A0A164LXC1 A0A310SLE7 G4VPT4 E2C4W7 A0A1Q3G289 A0A183P0Z4 A0A095CC93 A0A0C9QE78 A0A2A3E547 V9KWY0 A0A0P5DQL5 A0A0P5B8X8 A0A3R7Q020 A0A0P5BTE6 A0A0N8A2W8 A0A0N8DFG3 F4W5Y6 A0A0G4GQI5 A0A0P6HC18 A0A0P6BX82 A0A0P5VHB0 A0A026WHA9 A0A0J7L2A7 B4GCY3 E2B1R2 V9L595 A0A3L8DDS0 A0A0P5AY58 A0A183SQC8 V4B3E1 Q15ER9 H3B4S5 A0A151XH67 A0A0P6GEQ7 A0A195DFK1 A0A088AQV6
A0A3S2P0T4 A0A0L7LVI7 D6WH75 A0A1Y1KKD3 J3JYB1 A0A0V0GB22 A0A224XPH0 T1ICR0 A0A023F201 A0A1W4WKL0 A0A146M735 A0A2J7PDD1 A0A2P8YN48 A0A0A9YH30 A0A0K8SEQ3 J9JJX0 A0A067RBQ3 A0A068WGV8 W6UH69 A0A068Y522 A0A1D2NEB4 A0A1S3HYC4 A0A074Z2X9 A0A2R7X1R6 A0A183ALL3 A0A2G8JQ22 A0A1B6M475 A0A1S8WV56 A0A1B6GEN1 T1J491 A0A1B6HNT8 A0A210QH13 A0A2S2NII4 Q5DI52 G7YR01 A0A0X3PYL8 A0A183RSA0 A0A0P4WDQ5 A0A3P6TSE9 A0A154PN31 A0A0X3PSH1 A0A3R7DGC2 A0A2H8TID1 A0A0R3TNC8 A0A183MYI8 A0A164LXC1 A0A310SLE7 G4VPT4 E2C4W7 A0A1Q3G289 A0A183P0Z4 A0A095CC93 A0A0C9QE78 A0A2A3E547 V9KWY0 A0A0P5DQL5 A0A0P5B8X8 A0A3R7Q020 A0A0P5BTE6 A0A0N8A2W8 A0A0N8DFG3 F4W5Y6 A0A0G4GQI5 A0A0P6HC18 A0A0P6BX82 A0A0P5VHB0 A0A026WHA9 A0A0J7L2A7 B4GCY3 E2B1R2 V9L595 A0A3L8DDS0 A0A0P5AY58 A0A183SQC8 V4B3E1 Q15ER9 H3B4S5 A0A151XH67 A0A0P6GEQ7 A0A195DFK1 A0A088AQV6
Ontologies
PANTHER
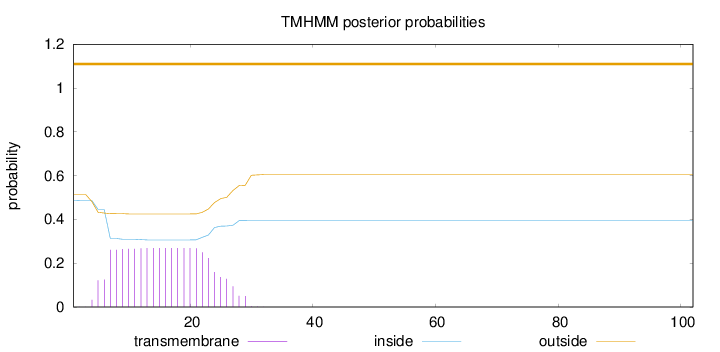
Topology
SignalP
Position: 1 - 29,
Likelihood: 0.967525
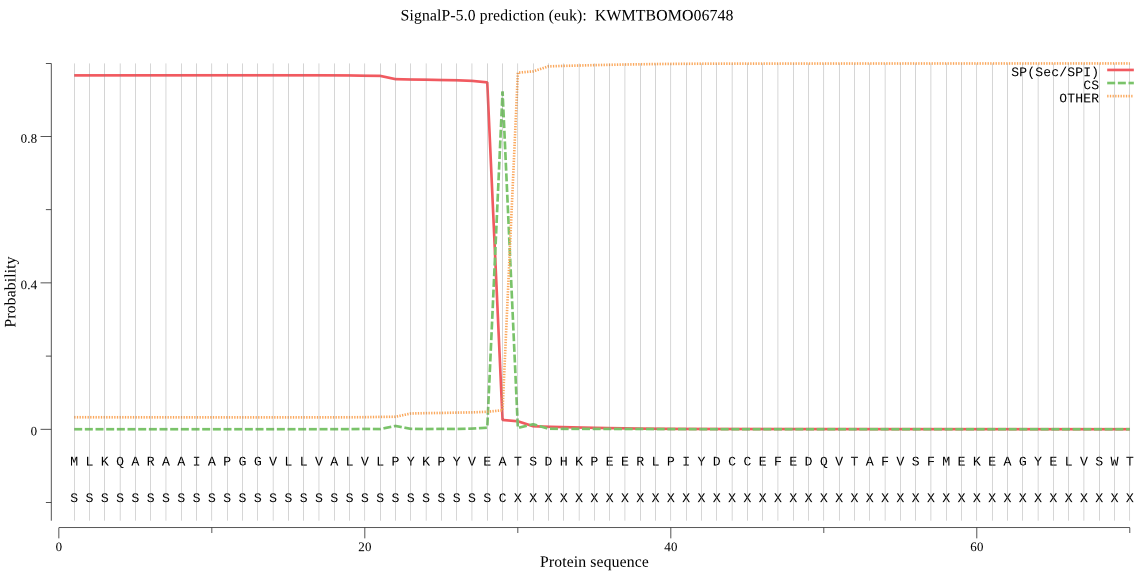
Length:
102
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.38242999999999
Exp number, first 60 AAs:
5.38061
Total prob of N-in:
0.48634
outside
1 - 102
Population Genetic Test Statistics
Pi
154.227005
Theta
140.354047
Tajima's D
0.495733
CLR
25.593992
CSRT
0.511924403779811
Interpretation
Uncertain
