Gene
KWMTBOMO06744
Pre Gene Modal
BGIBMGA012132
Annotation
PREDICTED:_sodium-coupled_monocarboxylate_transporter_2-like_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.955
Sequence
CDS
ATGAGATTAGGTGATGTCTCCAAGCGGGCGGTGTTGTGTCACATAACCAGCATCACTTTGCTCGGTGTGCCAGTCGAGATATATCTAAGAGGAACGCAATATTGGGCGTCTGCTCTGTCTCTGATCATTGTAACATTTCTCACAGCCGTTATTTATTTACCAGTGTTCCACAAACTGCAACTGTCGTCGAGCTTCGAGTATTTAGAGATAAGATTTTCGACACACACCAGAACGATTGCTGCCGTGATGTTCATCATCAGCAAATTGATGTTGCTGCCTATTGTTCCTTACGTGCCTCTCACAGCCTTCAGACTCGTAACAGATCAATTAGCCGAACGCATTACGGCTGTAGTATGCGTGTCTTGTGCTACTTTAATAGCTGTCGGTGGACTACAGGCTATAGTTACGCTCGGCATGGTCACAGTATTCCTGGCAATAGCCGGTACTGGCTTGCCGAGTGGATTAGCTCTTTTGCCAATGGGCTTGGCGAAGATGTGGGAACTTAATAGTCTCGGCGGGAGATTGGTTTTGTACGATCCCGATCCCGAATTGGAGCACCACACGTCCTTCTTTGCAGTCACATTAGCATTAAGTACCAATTGGCTGTGGAAAATGGCCTTGAGTCAGGGAACTCTCCAGAAGCTCCAAGCAGTATCGACGATAAATAAAGCAAGAGTATGTTTGATCATTGCATGCGTTGGAGTGATCTTAATGAAATTGCTTTCTTGTACACTCGGCTTGGCGCTGTACGCCTGGTTTGTCGGCTGCGATCCATTGCTCGCTGGAAAAATTAAGAAACATGAACAGGTAATGATGTTAAGTTAA
Protein
MRLGDVSKRAVLCHITSITLLGVPVEIYLRGTQYWASALSLIIVTFLTAVIYLPVFHKLQLSSSFEYLEIRFSTHTRTIAAVMFIISKLMLLPIVPYVPLTAFRLVTDQLAERITAVVCVSCATLIAVGGLQAIVTLGMVTVFLAIAGTGLPSGLALLPMGLAKMWELNSLGGRLVLYDPDPELEHHTSFFAVTLALSTNWLWKMALSQGTLQKLQAVSTINKARVCLIIACVGVILMKLLSCTLGLALYAWFVGCDPLLAGKIKKHEQVMMLS
Summary
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Uniprot
A0A194PEC1
A0A0N1IGB4
A0A2H1WJN4
A0A212F725
A0A0L7L199
A0A1Q3FQF7
+ More
B0W4K4 A0A026X377 E1ZVB7 A0A3L8DV00 A0A1B6I1F7 Q17K34 A0A1S4EZU6 A0A0J7KL16 A0A195C9N2 A0A067QTT6 E9IH55 A0A3S2LA31 A0A1B6EZ72 A0A151I4U6 A0A2J7QVC9 A0A182MC86 Q17K33 A0A0C9RWR2 A0A151JAS5 A0A195FKF8 A0A1B6F2A5 U5ESV6 A0A182K0S3 A0A067REM0 A0A1B6GEC2 D6WH77 A0A151WSC9 A0A139WI69 A0A1J1I490 A0A2P8ZE91 A0A182JI64 F4W533 A0A195CLV6 A0A195BP29 A0A158NDK2 F4WU94 A0A034VDC7 A0A034VEH2 A0A1B6HHC2 A0A1B6MFY6 A0A2H1VGG9 A0A195F3M3 A0A026W5H4 A0A182QV60 A0A1L8DER7 A0A212FHI2 A0A1J1ID30 A0A1B6CVL2 E2AQQ7 A0A170YED1 A0A0K8V9L1 A0A0K8VS11 W5JJX5 A0A1L8DES7 A0A194R0I5 A0A182F4T6 A0A1L8DES8 A0A232EG30 A0A154PA03 A0A195D9S5 A0A1B6D5M6 A0A084W5W1 A0A1L8DES4 A0A1B0CA85 A0A2J7PCD0 A0A1Y1L5S2 A0A2J7PCD1 E2AWG6 A0A0L7RCU1 A0A0K8URW5 A0A151XEF5 W8AJH7 W8ARK5 B4NAX6 A0A0A9WF43 A0A1B6EAY6 A0A182RDP6 A0A182NTY1 A0A182YR73 A0A182PLP8 A0A182NU01 T1IEF0 A0A182UC24 A0A232F205 W5JPR7 A0A3L8DFP4 A0A1B6HRF8 A0A1B6HG49 A0A026X262 A0A1B6JUL2 A0A1S4EZY9 Q17K36
B0W4K4 A0A026X377 E1ZVB7 A0A3L8DV00 A0A1B6I1F7 Q17K34 A0A1S4EZU6 A0A0J7KL16 A0A195C9N2 A0A067QTT6 E9IH55 A0A3S2LA31 A0A1B6EZ72 A0A151I4U6 A0A2J7QVC9 A0A182MC86 Q17K33 A0A0C9RWR2 A0A151JAS5 A0A195FKF8 A0A1B6F2A5 U5ESV6 A0A182K0S3 A0A067REM0 A0A1B6GEC2 D6WH77 A0A151WSC9 A0A139WI69 A0A1J1I490 A0A2P8ZE91 A0A182JI64 F4W533 A0A195CLV6 A0A195BP29 A0A158NDK2 F4WU94 A0A034VDC7 A0A034VEH2 A0A1B6HHC2 A0A1B6MFY6 A0A2H1VGG9 A0A195F3M3 A0A026W5H4 A0A182QV60 A0A1L8DER7 A0A212FHI2 A0A1J1ID30 A0A1B6CVL2 E2AQQ7 A0A170YED1 A0A0K8V9L1 A0A0K8VS11 W5JJX5 A0A1L8DES7 A0A194R0I5 A0A182F4T6 A0A1L8DES8 A0A232EG30 A0A154PA03 A0A195D9S5 A0A1B6D5M6 A0A084W5W1 A0A1L8DES4 A0A1B0CA85 A0A2J7PCD0 A0A1Y1L5S2 A0A2J7PCD1 E2AWG6 A0A0L7RCU1 A0A0K8URW5 A0A151XEF5 W8AJH7 W8ARK5 B4NAX6 A0A0A9WF43 A0A1B6EAY6 A0A182RDP6 A0A182NTY1 A0A182YR73 A0A182PLP8 A0A182NU01 T1IEF0 A0A182UC24 A0A232F205 W5JPR7 A0A3L8DFP4 A0A1B6HRF8 A0A1B6HG49 A0A026X262 A0A1B6JUL2 A0A1S4EZY9 Q17K36
Pubmed
EMBL
KQ459606
KPI91701.1
KQ460499
KPJ14275.1
ODYU01009109
SOQ53295.1
+ More
AGBW02009946 OWR49524.1 JTDY01003605 KOB69273.1 GFDL01005382 JAV29663.1 DS231838 EDS33849.1 KK107020 EZA62471.1 GL434491 EFN74893.1 QOIP01000004 RLU23719.1 GECU01026947 JAS80759.1 CH477228 EAT47058.1 LBMM01006054 KMQ90977.1 KQ978143 KYM96823.1 KK853282 KDR09026.1 GL763174 EFZ20108.1 RSAL01000346 RVE42323.1 GECZ01026618 JAS43151.1 KQ976445 KYM86132.1 NEVH01010477 PNF32536.1 AXCM01008724 EAT47059.1 GBYB01013490 JAG83257.1 KQ979275 KYN22033.1 KQ981522 KYN40747.1 GECZ01025434 JAS44335.1 GANO01003055 JAB56816.1 KK852746 KDR17314.1 GECZ01009116 JAS60653.1 KQ971330 EFA00982.1 KQ982775 KYQ50770.1 KQ971342 KYB27604.1 CVRI01000038 CRK94412.1 PYGN01000082 PSN54824.1 GL887596 EGI70729.1 KQ977600 KYN01452.1 KQ976424 KYM88272.1 ADTU01012644 GL888355 EGI62202.1 GAKP01018442 GAKP01018438 JAC40514.1 GAKP01018445 GAKP01018440 JAC40507.1 GECU01033701 JAS74005.1 GEBQ01005155 JAT34822.1 ODYU01002426 SOQ39896.1 KQ981820 KYN35180.1 KK107399 QOIP01000002 EZA51327.1 RLU25917.1 AXCN02000807 GFDF01009132 JAV04952.1 AGBW02008503 OWR53185.1 CVRI01000047 CRK98115.1 GEDC01019897 JAS17401.1 GL441804 EFN64246.1 GEMB01003380 JAR99834.1 GDHF01017084 GDHF01007157 JAI35230.1 JAI45157.1 GDHF01023020 GDHF01010647 JAI29294.1 JAI41667.1 ADMH02001219 ETN63613.1 GFDF01009121 JAV04963.1 KQ461108 KPJ09351.1 GFDF01009133 JAV04951.1 NNAY01004898 OXU17272.1 KQ434856 KZC08682.1 KQ981082 KYN09670.1 GEDC01016316 JAS20982.1 ATLV01020702 KE525305 KFB45605.1 GFDF01009122 JAV04962.1 AJWK01003402 NEVH01027063 PNF13986.1 GEZM01068536 JAV66906.1 PNF13988.1 GL443286 EFN62266.1 KQ414614 KOC68807.1 GDHF01023059 GDHF01022877 JAI29255.1 JAI29437.1 KQ982254 KYQ58745.1 GAMC01017855 JAB88700.1 GAMC01017858 GAMC01017854 JAB88697.1 CH964232 EDW80940.1 GBHO01037185 JAG06419.1 GEDC01002200 JAS35098.1 ACPB03014081 NNAY01001288 OXU24478.1 ADMH02000463 ETN66382.1 QOIP01000008 RLU19285.1 GECU01030439 JAS77267.1 GECU01034036 JAS73670.1 KK107031 EZA62128.1 GECU01004840 JAT02867.1 EAT47056.1
AGBW02009946 OWR49524.1 JTDY01003605 KOB69273.1 GFDL01005382 JAV29663.1 DS231838 EDS33849.1 KK107020 EZA62471.1 GL434491 EFN74893.1 QOIP01000004 RLU23719.1 GECU01026947 JAS80759.1 CH477228 EAT47058.1 LBMM01006054 KMQ90977.1 KQ978143 KYM96823.1 KK853282 KDR09026.1 GL763174 EFZ20108.1 RSAL01000346 RVE42323.1 GECZ01026618 JAS43151.1 KQ976445 KYM86132.1 NEVH01010477 PNF32536.1 AXCM01008724 EAT47059.1 GBYB01013490 JAG83257.1 KQ979275 KYN22033.1 KQ981522 KYN40747.1 GECZ01025434 JAS44335.1 GANO01003055 JAB56816.1 KK852746 KDR17314.1 GECZ01009116 JAS60653.1 KQ971330 EFA00982.1 KQ982775 KYQ50770.1 KQ971342 KYB27604.1 CVRI01000038 CRK94412.1 PYGN01000082 PSN54824.1 GL887596 EGI70729.1 KQ977600 KYN01452.1 KQ976424 KYM88272.1 ADTU01012644 GL888355 EGI62202.1 GAKP01018442 GAKP01018438 JAC40514.1 GAKP01018445 GAKP01018440 JAC40507.1 GECU01033701 JAS74005.1 GEBQ01005155 JAT34822.1 ODYU01002426 SOQ39896.1 KQ981820 KYN35180.1 KK107399 QOIP01000002 EZA51327.1 RLU25917.1 AXCN02000807 GFDF01009132 JAV04952.1 AGBW02008503 OWR53185.1 CVRI01000047 CRK98115.1 GEDC01019897 JAS17401.1 GL441804 EFN64246.1 GEMB01003380 JAR99834.1 GDHF01017084 GDHF01007157 JAI35230.1 JAI45157.1 GDHF01023020 GDHF01010647 JAI29294.1 JAI41667.1 ADMH02001219 ETN63613.1 GFDF01009121 JAV04963.1 KQ461108 KPJ09351.1 GFDF01009133 JAV04951.1 NNAY01004898 OXU17272.1 KQ434856 KZC08682.1 KQ981082 KYN09670.1 GEDC01016316 JAS20982.1 ATLV01020702 KE525305 KFB45605.1 GFDF01009122 JAV04962.1 AJWK01003402 NEVH01027063 PNF13986.1 GEZM01068536 JAV66906.1 PNF13988.1 GL443286 EFN62266.1 KQ414614 KOC68807.1 GDHF01023059 GDHF01022877 JAI29255.1 JAI29437.1 KQ982254 KYQ58745.1 GAMC01017855 JAB88700.1 GAMC01017858 GAMC01017854 JAB88697.1 CH964232 EDW80940.1 GBHO01037185 JAG06419.1 GEDC01002200 JAS35098.1 ACPB03014081 NNAY01001288 OXU24478.1 ADMH02000463 ETN66382.1 QOIP01000008 RLU19285.1 GECU01030439 JAS77267.1 GECU01034036 JAS73670.1 KK107031 EZA62128.1 GECU01004840 JAT02867.1 EAT47056.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000037510
UP000002320
UP000053097
+ More
UP000000311 UP000279307 UP000008820 UP000036403 UP000078542 UP000027135 UP000283053 UP000078540 UP000235965 UP000075883 UP000078492 UP000078541 UP000075881 UP000007266 UP000075809 UP000183832 UP000245037 UP000075880 UP000007755 UP000005205 UP000075886 UP000000673 UP000069272 UP000215335 UP000076502 UP000030765 UP000092461 UP000053825 UP000007798 UP000075900 UP000075884 UP000076408 UP000075885 UP000015103 UP000075902
UP000000311 UP000279307 UP000008820 UP000036403 UP000078542 UP000027135 UP000283053 UP000078540 UP000235965 UP000075883 UP000078492 UP000078541 UP000075881 UP000007266 UP000075809 UP000183832 UP000245037 UP000075880 UP000007755 UP000005205 UP000075886 UP000000673 UP000069272 UP000215335 UP000076502 UP000030765 UP000092461 UP000053825 UP000007798 UP000075900 UP000075884 UP000076408 UP000075885 UP000015103 UP000075902
Interpro
IPR038377
Na/Glc_symporter_sf
+ More
IPR001734 Na/solute_symporter
IPR016024 ARM-type_fold
IPR020616 Thiolase_N
IPR020615 Thiolase_acyl_enz_int_AS
IPR016039 Thiolase-like
IPR020617 Thiolase_C
IPR018212 Na/solute_symporter_CS
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR007110 Ig-like_dom
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001734 Na/solute_symporter
IPR016024 ARM-type_fold
IPR020616 Thiolase_N
IPR020615 Thiolase_acyl_enz_int_AS
IPR016039 Thiolase-like
IPR020617 Thiolase_C
IPR018212 Na/solute_symporter_CS
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR007110 Ig-like_dom
IPR023298 ATPase_P-typ_TM_dom_sf
Gene 3D
ProteinModelPortal
A0A194PEC1
A0A0N1IGB4
A0A2H1WJN4
A0A212F725
A0A0L7L199
A0A1Q3FQF7
+ More
B0W4K4 A0A026X377 E1ZVB7 A0A3L8DV00 A0A1B6I1F7 Q17K34 A0A1S4EZU6 A0A0J7KL16 A0A195C9N2 A0A067QTT6 E9IH55 A0A3S2LA31 A0A1B6EZ72 A0A151I4U6 A0A2J7QVC9 A0A182MC86 Q17K33 A0A0C9RWR2 A0A151JAS5 A0A195FKF8 A0A1B6F2A5 U5ESV6 A0A182K0S3 A0A067REM0 A0A1B6GEC2 D6WH77 A0A151WSC9 A0A139WI69 A0A1J1I490 A0A2P8ZE91 A0A182JI64 F4W533 A0A195CLV6 A0A195BP29 A0A158NDK2 F4WU94 A0A034VDC7 A0A034VEH2 A0A1B6HHC2 A0A1B6MFY6 A0A2H1VGG9 A0A195F3M3 A0A026W5H4 A0A182QV60 A0A1L8DER7 A0A212FHI2 A0A1J1ID30 A0A1B6CVL2 E2AQQ7 A0A170YED1 A0A0K8V9L1 A0A0K8VS11 W5JJX5 A0A1L8DES7 A0A194R0I5 A0A182F4T6 A0A1L8DES8 A0A232EG30 A0A154PA03 A0A195D9S5 A0A1B6D5M6 A0A084W5W1 A0A1L8DES4 A0A1B0CA85 A0A2J7PCD0 A0A1Y1L5S2 A0A2J7PCD1 E2AWG6 A0A0L7RCU1 A0A0K8URW5 A0A151XEF5 W8AJH7 W8ARK5 B4NAX6 A0A0A9WF43 A0A1B6EAY6 A0A182RDP6 A0A182NTY1 A0A182YR73 A0A182PLP8 A0A182NU01 T1IEF0 A0A182UC24 A0A232F205 W5JPR7 A0A3L8DFP4 A0A1B6HRF8 A0A1B6HG49 A0A026X262 A0A1B6JUL2 A0A1S4EZY9 Q17K36
B0W4K4 A0A026X377 E1ZVB7 A0A3L8DV00 A0A1B6I1F7 Q17K34 A0A1S4EZU6 A0A0J7KL16 A0A195C9N2 A0A067QTT6 E9IH55 A0A3S2LA31 A0A1B6EZ72 A0A151I4U6 A0A2J7QVC9 A0A182MC86 Q17K33 A0A0C9RWR2 A0A151JAS5 A0A195FKF8 A0A1B6F2A5 U5ESV6 A0A182K0S3 A0A067REM0 A0A1B6GEC2 D6WH77 A0A151WSC9 A0A139WI69 A0A1J1I490 A0A2P8ZE91 A0A182JI64 F4W533 A0A195CLV6 A0A195BP29 A0A158NDK2 F4WU94 A0A034VDC7 A0A034VEH2 A0A1B6HHC2 A0A1B6MFY6 A0A2H1VGG9 A0A195F3M3 A0A026W5H4 A0A182QV60 A0A1L8DER7 A0A212FHI2 A0A1J1ID30 A0A1B6CVL2 E2AQQ7 A0A170YED1 A0A0K8V9L1 A0A0K8VS11 W5JJX5 A0A1L8DES7 A0A194R0I5 A0A182F4T6 A0A1L8DES8 A0A232EG30 A0A154PA03 A0A195D9S5 A0A1B6D5M6 A0A084W5W1 A0A1L8DES4 A0A1B0CA85 A0A2J7PCD0 A0A1Y1L5S2 A0A2J7PCD1 E2AWG6 A0A0L7RCU1 A0A0K8URW5 A0A151XEF5 W8AJH7 W8ARK5 B4NAX6 A0A0A9WF43 A0A1B6EAY6 A0A182RDP6 A0A182NTY1 A0A182YR73 A0A182PLP8 A0A182NU01 T1IEF0 A0A182UC24 A0A232F205 W5JPR7 A0A3L8DFP4 A0A1B6HRF8 A0A1B6HG49 A0A026X262 A0A1B6JUL2 A0A1S4EZY9 Q17K36
PDB
5NVA
E-value=1.64193e-07,
Score=131
Ontologies
GO
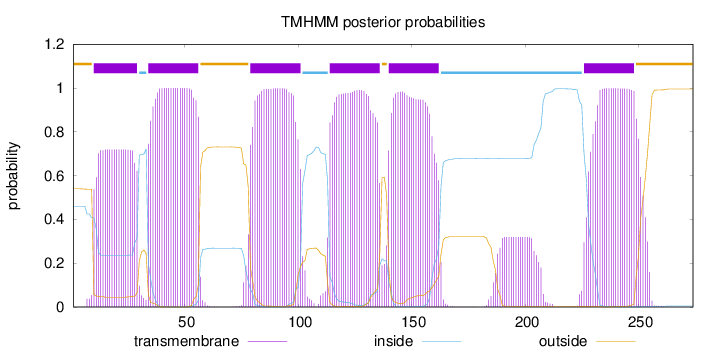
Topology
Length:
274
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
133.38698
Exp number, first 60 AAs:
36.62254
Total prob of N-in:
0.46040
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 29
inside
30 - 33
TMhelix
34 - 56
outside
57 - 78
TMhelix
79 - 101
inside
102 - 113
TMhelix
114 - 136
outside
137 - 139
TMhelix
140 - 162
inside
163 - 225
TMhelix
226 - 248
outside
249 - 274
Population Genetic Test Statistics
Pi
214.844805
Theta
163.406862
Tajima's D
1.206481
CLR
0
CSRT
0.717514124293785
Interpretation
Uncertain
