Gene
KWMTBOMO06743
Pre Gene Modal
BGIBMGA012132
Annotation
Sodium-coupled_monocarboxylate_transporter_2_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.819
Sequence
CDS
ATGTTTCTATTCTCACCTGCCGTTTTAGTTCCGCAATTCTTGAACCACCTGTCGACAATGTTTCCTGGTATCTGTGGTATATTCATTATATCGATATTCAGCGCCACTGCCGGGTGCATAGCAACGATAATTAATTCAATAACTGGAGTGTTGTTTGAGGAATTCATCAGGCCGTGGATGCCGCAAGACACCAACGAACTAACATGTTGCAAAATCATGAAAGTTCTTTGTATGCTGGTTGGGTGTTACTGCGCATGCGTGGTCTGGTTTGCGAAGGACTTAGACCGCTTGCAGCACGTCGCTTCAGGTGTGACAGGAGTATCTGCAGGCACTTTACTAGGCTTGTTCACGCTTGGCATTGTCTTCTCGAGGGCAAACAGTTGTGGTGCATTAGGAGGGAGCTTGCTAAGTCTACTCTTATGTGGATGGCTGCTCATTGGCGCGGAGAATTCTCTAGCAAGAGGTGCATTAACGTTCCAAGGAAAACCTTTGACAACCGCTGGCTGTGGCAAAGTCAACTTCACGCTCACCGCCTTTGATACCACTACAACTCCAATGCCTAATCTACTAAACCCAATCGCCAGCAAACCTCTTCAGTCACTCTTCAGGATATCATTCACGTACTGTCCGTTCGCTGGAGCAATAACGGTGTTGCTTATAGGCGTACCCCTAAGTTATATGACCGGAAAGTCTCGCACCGATTCCATGAACCCTGACGTATTCTGCCCGCTGACCCAGAGCATGCTGCACAAGCCCCAGATGCGAAGCAGGACCGTTTCCGTGGAGGCCAGAGCTCCGAACACTCAGGAAGTGTATTTAGCTTTGGACGAAGCTCTCAAGCACCTAAAGGATGTCATTGATCGATAA
Protein
MFLFSPAVLVPQFLNHLSTMFPGICGIFIISIFSATAGCIATIINSITGVLFEEFIRPWMPQDTNELTCCKIMKVLCMLVGCYCACVVWFAKDLDRLQHVASGVTGVSAGTLLGLFTLGIVFSRANSCGALGGSLLSLLLCGWLLIGAENSLARGALTFQGKPLTTAGCGKVNFTLTAFDTTTTPMPNLLNPIASKPLQSLFRISFTYCPFAGAITVLLIGVPLSYMTGKSRTDSMNPDVFCPLTQSMLHKPQMRSRTVSVEARAPNTQEVYLALDEALKHLKDVIDR
Summary
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Uniprot
A0A194PEC1
A0A2A4JNK8
A0A0N1IGB4
A0A2H1WJN4
A0A212F725
A0A1B6EH87
+ More
A0A1B6EC94 A0A2J7PCC6 A0A2J7PCD2 A0A2J7PCD1 A0A2J7PCD0 A0A0T6AVN4 A0A0M8ZM94 A0A154PA03 A0A1W4WGZ1 A0A2P8ZEB9 A0A0C9RWR2 A0A1B6L6J6 A0A2J7QVC9 A0A067REM0 A0A1Y1L4L3 A0A026X262 A0A3L8DFP4 A0A182VW47 E9IR19 A0A1Y1L5S2 E0VXB0 A0A151WGZ9 A0A195BZT4 A0A195BP29 A0A158NDK2 A0A182RSH4 A0A0J7N294 A0A151XEF5 A0A195FWJ9 E2AWG6 A0A195DCV9 A0A2A3E2H5 A0A067RC05 A0A2J7PJL8 A0A0A9WF43 A0A182FS71 A0A088AH95 A0A182SGA4 A0A2R7VQN2 A0A0J7L3T5 A0A2J7QVB3 A0A1B6K7H1 A0A1W4XIB1 A0A1B6G815 A0A2J7QVB8 W5J189 F4WJQ2 A0A195D9S5 A0A034VW29 A0A034VXQ3 A0A3S2LA31 D6WJW9 A0A1B6JU79 A0A034VYN0 A0A139WI69 A0A1B6EV65 E2BY05 A0A0K8WKR1 A0A195F3M3 A0A1B0DBD6 A0A0K8U1A2 A0A1B6LPJ9 A0A0L7LDB5 A0A182K1U7 A0A195CLV6 F4W533 A0A2J7QVB4 A0A1J1HTS9 A0A182PJ36 A0A158NS68 A0A195BBB9 T1HJF5 A0A182NQW1 A0A182QU12 A0A0L7RCU1 A0A1B0CT26 E2BD54 A0A1B6JH88 A0A182L8F2 A0A026W5H4 Q16SA5 A0A1S4FR06 A0A182IYN7 A0A1B6M092 A0A1B6D7D6 A0A1W4X8M1 A0A182YHT2 A0A182TWQ2 A0A0A1XEH3 A0A182VI42 Q175Y8
A0A1B6EC94 A0A2J7PCC6 A0A2J7PCD2 A0A2J7PCD1 A0A2J7PCD0 A0A0T6AVN4 A0A0M8ZM94 A0A154PA03 A0A1W4WGZ1 A0A2P8ZEB9 A0A0C9RWR2 A0A1B6L6J6 A0A2J7QVC9 A0A067REM0 A0A1Y1L4L3 A0A026X262 A0A3L8DFP4 A0A182VW47 E9IR19 A0A1Y1L5S2 E0VXB0 A0A151WGZ9 A0A195BZT4 A0A195BP29 A0A158NDK2 A0A182RSH4 A0A0J7N294 A0A151XEF5 A0A195FWJ9 E2AWG6 A0A195DCV9 A0A2A3E2H5 A0A067RC05 A0A2J7PJL8 A0A0A9WF43 A0A182FS71 A0A088AH95 A0A182SGA4 A0A2R7VQN2 A0A0J7L3T5 A0A2J7QVB3 A0A1B6K7H1 A0A1W4XIB1 A0A1B6G815 A0A2J7QVB8 W5J189 F4WJQ2 A0A195D9S5 A0A034VW29 A0A034VXQ3 A0A3S2LA31 D6WJW9 A0A1B6JU79 A0A034VYN0 A0A139WI69 A0A1B6EV65 E2BY05 A0A0K8WKR1 A0A195F3M3 A0A1B0DBD6 A0A0K8U1A2 A0A1B6LPJ9 A0A0L7LDB5 A0A182K1U7 A0A195CLV6 F4W533 A0A2J7QVB4 A0A1J1HTS9 A0A182PJ36 A0A158NS68 A0A195BBB9 T1HJF5 A0A182NQW1 A0A182QU12 A0A0L7RCU1 A0A1B0CT26 E2BD54 A0A1B6JH88 A0A182L8F2 A0A026W5H4 Q16SA5 A0A1S4FR06 A0A182IYN7 A0A1B6M092 A0A1B6D7D6 A0A1W4X8M1 A0A182YHT2 A0A182TWQ2 A0A0A1XEH3 A0A182VI42 Q175Y8
Pubmed
EMBL
KQ459606
KPI91701.1
NWSH01001031
PCG73023.1
KQ460499
KPJ14275.1
+ More
ODYU01009109 SOQ53295.1 AGBW02009946 OWR49524.1 GEDC01000002 JAS37296.1 GEDC01001788 JAS35510.1 NEVH01027063 PNF13989.1 PNF13987.1 PNF13988.1 PNF13986.1 LJIG01022728 KRT79021.1 KQ438551 KOX67190.1 KQ434856 KZC08682.1 PYGN01000082 PSN54836.1 GBYB01013490 JAG83257.1 GEBQ01020648 JAT19329.1 NEVH01010477 PNF32536.1 KK852746 KDR17314.1 GEZM01068537 JAV66905.1 KK107031 EZA62128.1 QOIP01000008 RLU19285.1 GL765043 EFZ16973.1 GEZM01068536 JAV66906.1 DS235830 EEB18016.1 KQ983136 KYQ47116.1 KQ978457 KYM93850.1 KQ976424 KYM88272.1 ADTU01012644 LBMM01011538 KMQ86775.1 KQ982254 KYQ58745.1 KQ981208 KYN44687.1 GL443286 EFN62266.1 KQ980989 KYN10745.1 KZ288427 PBC25900.1 KDR17310.1 NEVH01024946 PNF16521.1 GBHO01037185 JAG06419.1 KK854027 PTY09716.1 LBMM01000877 KMQ97311.1 PNF32528.1 GECU01000325 JAT07382.1 GECZ01011177 JAS58592.1 PNF32527.1 ADMH02002189 ETN57897.1 GL888186 EGI65544.1 KQ981082 KYN09670.1 GAKP01012303 JAC46649.1 GAKP01012307 JAC46645.1 RSAL01000346 RVE42323.1 KQ971342 EFA03651.2 GECU01004941 JAT02766.1 GAKP01012309 JAC46643.1 KYB27604.1 GECZ01027950 JAS41819.1 GL451363 EFN79439.1 GDHF01000591 JAI51723.1 KQ981820 KYN35180.1 AJVK01004852 GDHF01032026 JAI20288.1 GEBQ01014345 JAT25632.1 JTDY01001579 KOB73472.1 KQ977600 KYN01452.1 GL887596 EGI70729.1 PNF32525.1 CVRI01000021 CRK91471.1 ADTU01024524 ADTU01024525 KQ976528 KYM81828.1 ACPB03021235 AXCN02001489 KQ414614 KOC68807.1 AJWK01026951 AJWK01026952 AJWK01026953 AJWK01026954 GL447569 EFN86362.1 GECU01009177 JAS98529.1 KK107399 QOIP01000002 EZA51327.1 RLU25917.1 CH477679 EAT37341.1 GEBQ01010667 JAT29310.1 GEDC01015697 JAS21601.1 GBXI01005399 JAD08893.1 CH477393 EAT41908.1
ODYU01009109 SOQ53295.1 AGBW02009946 OWR49524.1 GEDC01000002 JAS37296.1 GEDC01001788 JAS35510.1 NEVH01027063 PNF13989.1 PNF13987.1 PNF13988.1 PNF13986.1 LJIG01022728 KRT79021.1 KQ438551 KOX67190.1 KQ434856 KZC08682.1 PYGN01000082 PSN54836.1 GBYB01013490 JAG83257.1 GEBQ01020648 JAT19329.1 NEVH01010477 PNF32536.1 KK852746 KDR17314.1 GEZM01068537 JAV66905.1 KK107031 EZA62128.1 QOIP01000008 RLU19285.1 GL765043 EFZ16973.1 GEZM01068536 JAV66906.1 DS235830 EEB18016.1 KQ983136 KYQ47116.1 KQ978457 KYM93850.1 KQ976424 KYM88272.1 ADTU01012644 LBMM01011538 KMQ86775.1 KQ982254 KYQ58745.1 KQ981208 KYN44687.1 GL443286 EFN62266.1 KQ980989 KYN10745.1 KZ288427 PBC25900.1 KDR17310.1 NEVH01024946 PNF16521.1 GBHO01037185 JAG06419.1 KK854027 PTY09716.1 LBMM01000877 KMQ97311.1 PNF32528.1 GECU01000325 JAT07382.1 GECZ01011177 JAS58592.1 PNF32527.1 ADMH02002189 ETN57897.1 GL888186 EGI65544.1 KQ981082 KYN09670.1 GAKP01012303 JAC46649.1 GAKP01012307 JAC46645.1 RSAL01000346 RVE42323.1 KQ971342 EFA03651.2 GECU01004941 JAT02766.1 GAKP01012309 JAC46643.1 KYB27604.1 GECZ01027950 JAS41819.1 GL451363 EFN79439.1 GDHF01000591 JAI51723.1 KQ981820 KYN35180.1 AJVK01004852 GDHF01032026 JAI20288.1 GEBQ01014345 JAT25632.1 JTDY01001579 KOB73472.1 KQ977600 KYN01452.1 GL887596 EGI70729.1 PNF32525.1 CVRI01000021 CRK91471.1 ADTU01024524 ADTU01024525 KQ976528 KYM81828.1 ACPB03021235 AXCN02001489 KQ414614 KOC68807.1 AJWK01026951 AJWK01026952 AJWK01026953 AJWK01026954 GL447569 EFN86362.1 GECU01009177 JAS98529.1 KK107399 QOIP01000002 EZA51327.1 RLU25917.1 CH477679 EAT37341.1 GEBQ01010667 JAT29310.1 GEDC01015697 JAS21601.1 GBXI01005399 JAD08893.1 CH477393 EAT41908.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000007151
UP000235965
UP000053105
+ More
UP000076502 UP000192223 UP000245037 UP000027135 UP000053097 UP000279307 UP000075920 UP000009046 UP000075809 UP000078542 UP000078540 UP000005205 UP000075900 UP000036403 UP000078541 UP000000311 UP000078492 UP000242457 UP000069272 UP000005203 UP000075901 UP000000673 UP000007755 UP000283053 UP000007266 UP000008237 UP000092462 UP000037510 UP000075881 UP000183832 UP000075885 UP000015103 UP000075884 UP000075886 UP000053825 UP000092461 UP000075882 UP000008820 UP000075880 UP000076408 UP000075902 UP000075903
UP000076502 UP000192223 UP000245037 UP000027135 UP000053097 UP000279307 UP000075920 UP000009046 UP000075809 UP000078542 UP000078540 UP000005205 UP000075900 UP000036403 UP000078541 UP000000311 UP000078492 UP000242457 UP000069272 UP000005203 UP000075901 UP000000673 UP000007755 UP000283053 UP000007266 UP000008237 UP000092462 UP000037510 UP000075881 UP000183832 UP000075885 UP000015103 UP000075884 UP000075886 UP000053825 UP000092461 UP000075882 UP000008820 UP000075880 UP000076408 UP000075902 UP000075903
Pfam
PF00474 SSF
Interpro
SUPFAM
SSF81665
SSF81665
Gene 3D
ProteinModelPortal
A0A194PEC1
A0A2A4JNK8
A0A0N1IGB4
A0A2H1WJN4
A0A212F725
A0A1B6EH87
+ More
A0A1B6EC94 A0A2J7PCC6 A0A2J7PCD2 A0A2J7PCD1 A0A2J7PCD0 A0A0T6AVN4 A0A0M8ZM94 A0A154PA03 A0A1W4WGZ1 A0A2P8ZEB9 A0A0C9RWR2 A0A1B6L6J6 A0A2J7QVC9 A0A067REM0 A0A1Y1L4L3 A0A026X262 A0A3L8DFP4 A0A182VW47 E9IR19 A0A1Y1L5S2 E0VXB0 A0A151WGZ9 A0A195BZT4 A0A195BP29 A0A158NDK2 A0A182RSH4 A0A0J7N294 A0A151XEF5 A0A195FWJ9 E2AWG6 A0A195DCV9 A0A2A3E2H5 A0A067RC05 A0A2J7PJL8 A0A0A9WF43 A0A182FS71 A0A088AH95 A0A182SGA4 A0A2R7VQN2 A0A0J7L3T5 A0A2J7QVB3 A0A1B6K7H1 A0A1W4XIB1 A0A1B6G815 A0A2J7QVB8 W5J189 F4WJQ2 A0A195D9S5 A0A034VW29 A0A034VXQ3 A0A3S2LA31 D6WJW9 A0A1B6JU79 A0A034VYN0 A0A139WI69 A0A1B6EV65 E2BY05 A0A0K8WKR1 A0A195F3M3 A0A1B0DBD6 A0A0K8U1A2 A0A1B6LPJ9 A0A0L7LDB5 A0A182K1U7 A0A195CLV6 F4W533 A0A2J7QVB4 A0A1J1HTS9 A0A182PJ36 A0A158NS68 A0A195BBB9 T1HJF5 A0A182NQW1 A0A182QU12 A0A0L7RCU1 A0A1B0CT26 E2BD54 A0A1B6JH88 A0A182L8F2 A0A026W5H4 Q16SA5 A0A1S4FR06 A0A182IYN7 A0A1B6M092 A0A1B6D7D6 A0A1W4X8M1 A0A182YHT2 A0A182TWQ2 A0A0A1XEH3 A0A182VI42 Q175Y8
A0A1B6EC94 A0A2J7PCC6 A0A2J7PCD2 A0A2J7PCD1 A0A2J7PCD0 A0A0T6AVN4 A0A0M8ZM94 A0A154PA03 A0A1W4WGZ1 A0A2P8ZEB9 A0A0C9RWR2 A0A1B6L6J6 A0A2J7QVC9 A0A067REM0 A0A1Y1L4L3 A0A026X262 A0A3L8DFP4 A0A182VW47 E9IR19 A0A1Y1L5S2 E0VXB0 A0A151WGZ9 A0A195BZT4 A0A195BP29 A0A158NDK2 A0A182RSH4 A0A0J7N294 A0A151XEF5 A0A195FWJ9 E2AWG6 A0A195DCV9 A0A2A3E2H5 A0A067RC05 A0A2J7PJL8 A0A0A9WF43 A0A182FS71 A0A088AH95 A0A182SGA4 A0A2R7VQN2 A0A0J7L3T5 A0A2J7QVB3 A0A1B6K7H1 A0A1W4XIB1 A0A1B6G815 A0A2J7QVB8 W5J189 F4WJQ2 A0A195D9S5 A0A034VW29 A0A034VXQ3 A0A3S2LA31 D6WJW9 A0A1B6JU79 A0A034VYN0 A0A139WI69 A0A1B6EV65 E2BY05 A0A0K8WKR1 A0A195F3M3 A0A1B0DBD6 A0A0K8U1A2 A0A1B6LPJ9 A0A0L7LDB5 A0A182K1U7 A0A195CLV6 F4W533 A0A2J7QVB4 A0A1J1HTS9 A0A182PJ36 A0A158NS68 A0A195BBB9 T1HJF5 A0A182NQW1 A0A182QU12 A0A0L7RCU1 A0A1B0CT26 E2BD54 A0A1B6JH88 A0A182L8F2 A0A026W5H4 Q16SA5 A0A1S4FR06 A0A182IYN7 A0A1B6M092 A0A1B6D7D6 A0A1W4X8M1 A0A182YHT2 A0A182TWQ2 A0A0A1XEH3 A0A182VI42 Q175Y8
Ontologies
GO
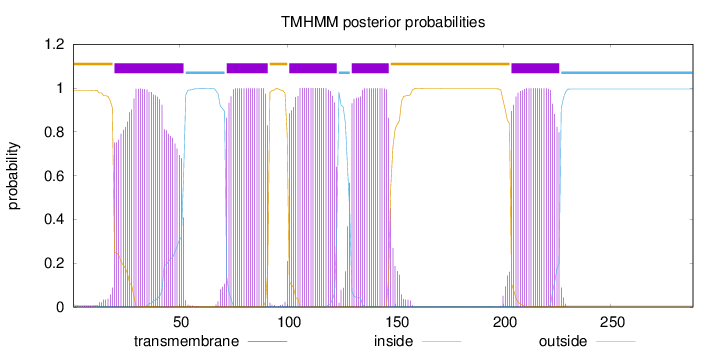
Topology
Length:
288
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.9263
Exp number, first 60 AAs:
28.59582
Total prob of N-in:
0.01006
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 52
inside
53 - 71
TMhelix
72 - 91
outside
92 - 100
TMhelix
101 - 123
inside
124 - 129
TMhelix
130 - 147
outside
148 - 203
TMhelix
204 - 226
inside
227 - 288
Population Genetic Test Statistics
Pi
227.800353
Theta
157.982173
Tajima's D
0.859851
CLR
0.17827
CSRT
0.624318784060797
Interpretation
Uncertain
