Gene
KWMTBOMO06740 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012134
Annotation
PREDICTED:_uncharacterized_protein_LOC101735671_isoform_X2_[Bombyx_mori]
Full name
E3 ubiquitin-protein ligase
Location in the cell
Nuclear Reliability : 2.282
Sequence
CDS
ATGTCGCAGAATCTGGTCACACAATCGCAGCTTACATTAGACTCGGAAAGTCGAAAGTCCGTAGTCCTGAATTCGCTACTGGAGACTGACCATTATGAGAGCATCATCAAAGACCTGACGTGCAAAAGATGCTCAAAATACATGAAGCCGCCTATTCACCTCTGCGTGGACGGACACAGTATCTGCGGTCCTTGTTATGACAAAAGCTATCAATGTCACGTGTGCAAAAAAGAATTTTCACCGATCCGTTCCGCCGTGCTGGAGTCGTTGGCAAACAAAGTCTTGTTCCCGTGCACGAATGCGGGATGCCCAAAACACGCCACGCTCCCGCTACTGGAGAAGCACACTCCGCACTGCCAGTTCAGAATCATAAACTGCTTTATGGCTAGAGTATACGGTGAGTGCACGTGGGAGGGGCGCGCGGGCGACTGGATGGACCACTGCTTCTCGGAACATAAACAAAAGGTCACTGACCTGCCCTTCATCACTGTCAAAGACAAATGGGACGCTAAGAGGACAGAGCCGATCCTAAACTATTTCCTGCTAAGATGCTTCGAAAAAGTTTTTAATGTTTACCAGATCTATGATAAAAGAGGAGGTCGAATGATGTGGACAGTGCTCGTAAACGATGACAATGCAGAGAAATTTTATTTCGAAATCGACATCTTCCTTCCAAATCTACCGAGCAAAAGGATCGTGTACCGACGCCCCTGTAAATGCGAGAAGGACGCTGATTTCCTGGAACACACACAGAACGTGTACATTCCCGTTGAAAACGTTTTCTCTATGCTCGATGACAGCGAAGCCTTGAATTTCATTGTTAGAATCGGTGAAGTAGAAAATTTACCTCTGCTCGATACACCGACAGATAGCGAGAGCCGTATTCTTCTGAACCACGAAGATCATAAATAA
Protein
MSQNLVTQSQLTLDSESRKSVVLNSLLETDHYESIIKDLTCKRCSKYMKPPIHLCVDGHSICGPCYDKSYQCHVCKKEFSPIRSAVLESLANKVLFPCTNAGCPKHATLPLLEKHTPHCQFRIINCFMARVYGECTWEGRAGDWMDHCFSEHKQKVTDLPFITVKDKWDAKRTEPILNYFLLRCFEKVFNVYQIYDKRGGRMMWTVLVNDDNAEKFYFEIDIFLPNLPSKRIVYRRPCKCEKDADFLEHTQNVYIPVENVFSMLDDSEALNFIVRIGEVENLPLLDTPTDSESRILLNHEDHK
Summary
Description
E3 ubiquitin-protein ligase that mediates ubiquitination and subsequent proteasomal degradation of target proteins. E3 ubiquitin ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates.
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Similarity
Belongs to the SINA (Seven in absentia) family.
Belongs to the eIF-2B alpha/beta/delta subunits family.
Belongs to the eIF-2B alpha/beta/delta subunits family.
Uniprot
H9JRH4
A0A194PFA0
A0A0N1IEK6
A0A212FC73
A0A2H1WI24
A0A2A4K4C3
+ More
A0A2A4K3L4 A0A2A4K3N6 A0A2A4K2L6 A0A1S4FBP6 Q178N6 A0A1S4G471 Q1DGT7 B0X888 A0A182M2G9 A0A182W5Q5 A0A182RHN8 A0A182R0S6 A0A084VQD2 A0A182J359 A0A182YCV2 A0A182UP81 A0A182N8I5 A0A182TUG8 A0A182FJ00 A0A182XJZ9 Q7QF99 A0A182IAX9 A0A1B0D8Y9 A0A1B0CHF1 T1PC13 A0A1I8ND94 A0A1I8Q1U1 B4L1M7 W8BDB3 A0A0M4EIM2 A0A1A9Z212 A0A1A9XZY6 A0A1A9VD56 A0A182KLC8 A0A0A1XRW6 A0A034V5G1 A0A034V863 Q29HR3 A0A1B0B403 B4GYG0 A0A0K8UQ93 A0A0K8UPG7 A0A1B0FAC1 A0A3B0JXW5 A0A0R3P4Z5 B4N1V3 B3MWG9 B4I9Y9 A0A0P9BV27 Q9W4W4 B4R3T9 B4MG75 B3NTW1 A0A1W4VF65 B4JL29 B4Q0X0 Q8MTX6 A0A1W4XAW8 A0A0R3P416 A0A139WEW0 A0A1Y1LLZ1 D6W9Y6 V5GSR3 O76862 D6WQJ0 A0A1W4XCK0 A0A1W4XCS1 A0A2J7QN57 A0A1W4WN84 A0A2P8Z4M8 A0A1E1WES9
A0A2A4K3L4 A0A2A4K3N6 A0A2A4K2L6 A0A1S4FBP6 Q178N6 A0A1S4G471 Q1DGT7 B0X888 A0A182M2G9 A0A182W5Q5 A0A182RHN8 A0A182R0S6 A0A084VQD2 A0A182J359 A0A182YCV2 A0A182UP81 A0A182N8I5 A0A182TUG8 A0A182FJ00 A0A182XJZ9 Q7QF99 A0A182IAX9 A0A1B0D8Y9 A0A1B0CHF1 T1PC13 A0A1I8ND94 A0A1I8Q1U1 B4L1M7 W8BDB3 A0A0M4EIM2 A0A1A9Z212 A0A1A9XZY6 A0A1A9VD56 A0A182KLC8 A0A0A1XRW6 A0A034V5G1 A0A034V863 Q29HR3 A0A1B0B403 B4GYG0 A0A0K8UQ93 A0A0K8UPG7 A0A1B0FAC1 A0A3B0JXW5 A0A0R3P4Z5 B4N1V3 B3MWG9 B4I9Y9 A0A0P9BV27 Q9W4W4 B4R3T9 B4MG75 B3NTW1 A0A1W4VF65 B4JL29 B4Q0X0 Q8MTX6 A0A1W4XAW8 A0A0R3P416 A0A139WEW0 A0A1Y1LLZ1 D6W9Y6 V5GSR3 O76862 D6WQJ0 A0A1W4XCK0 A0A1W4XCS1 A0A2J7QN57 A0A1W4WN84 A0A2P8Z4M8 A0A1E1WES9
EC Number
2.3.2.27
Pubmed
EMBL
BABH01016455
KQ459606
KPI91698.1
KQ460499
KPJ14278.1
AGBW02009213
+ More
OWR51330.1 ODYU01008479 SOQ52114.1 NWSH01000197 PCG78502.1 PCG78498.1 PCG78504.1 PCG78501.1 CH477361 EAT42654.1 CH900425 EAT32367.1 DS232478 EDS42357.1 AXCM01001073 AXCN02002231 ATLV01015189 KE525003 KFB40176.1 AAAB01008846 EAA06700.5 APCN01001358 AJVK01012846 AJWK01012309 AJWK01012310 KA646229 AFP60858.1 CH933810 EDW07663.2 GAMC01018788 GAMC01018786 JAB87769.1 CP012528 ALC48778.1 GBXI01000238 JAD14054.1 GAKP01021228 JAC37724.1 GAKP01021229 JAC37723.1 CH379064 EAL31695.1 JXJN01008138 JXJN01008139 CH479197 EDW27816.1 GDHF01023593 JAI28721.1 GDHF01023715 JAI28599.1 CCAG010006092 OUUW01000011 SPP86905.1 KRT06464.1 CH963925 EDW78342.1 CH902625 EDV34954.1 CH480825 EDW44020.1 KPU75371.1 AE014298 BT044595 AAF45811.2 ACI31295.1 CM000366 EDX16951.1 CH940672 EDW57398.2 CH954180 EDV45669.1 CH916370 EDW00282.1 CM000162 EDX01337.2 AY047979 AAL08361.1 KRT06463.1 KQ971354 KYB26391.1 GEZM01051860 GEZM01051859 GEZM01051858 GEZM01051857 JAV74659.1 KQ971312 EEZ98089.2 GALX01001247 JAB67219.1 AL023874 CAA19647.1 EFA06066.2 NEVH01013199 PNF30025.1 PYGN01000196 PSN51458.1 GDQN01005556 JAT85498.1
OWR51330.1 ODYU01008479 SOQ52114.1 NWSH01000197 PCG78502.1 PCG78498.1 PCG78504.1 PCG78501.1 CH477361 EAT42654.1 CH900425 EAT32367.1 DS232478 EDS42357.1 AXCM01001073 AXCN02002231 ATLV01015189 KE525003 KFB40176.1 AAAB01008846 EAA06700.5 APCN01001358 AJVK01012846 AJWK01012309 AJWK01012310 KA646229 AFP60858.1 CH933810 EDW07663.2 GAMC01018788 GAMC01018786 JAB87769.1 CP012528 ALC48778.1 GBXI01000238 JAD14054.1 GAKP01021228 JAC37724.1 GAKP01021229 JAC37723.1 CH379064 EAL31695.1 JXJN01008138 JXJN01008139 CH479197 EDW27816.1 GDHF01023593 JAI28721.1 GDHF01023715 JAI28599.1 CCAG010006092 OUUW01000011 SPP86905.1 KRT06464.1 CH963925 EDW78342.1 CH902625 EDV34954.1 CH480825 EDW44020.1 KPU75371.1 AE014298 BT044595 AAF45811.2 ACI31295.1 CM000366 EDX16951.1 CH940672 EDW57398.2 CH954180 EDV45669.1 CH916370 EDW00282.1 CM000162 EDX01337.2 AY047979 AAL08361.1 KRT06463.1 KQ971354 KYB26391.1 GEZM01051860 GEZM01051859 GEZM01051858 GEZM01051857 JAV74659.1 KQ971312 EEZ98089.2 GALX01001247 JAB67219.1 AL023874 CAA19647.1 EFA06066.2 NEVH01013199 PNF30025.1 PYGN01000196 PSN51458.1 GDQN01005556 JAT85498.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000008820
+ More
UP000002320 UP000075883 UP000075920 UP000075900 UP000075886 UP000030765 UP000075880 UP000076408 UP000075903 UP000075884 UP000075902 UP000069272 UP000076407 UP000007062 UP000075840 UP000092462 UP000092461 UP000095301 UP000095300 UP000009192 UP000092553 UP000092445 UP000092443 UP000078200 UP000075882 UP000001819 UP000092460 UP000008744 UP000092444 UP000268350 UP000007798 UP000007801 UP000001292 UP000000803 UP000000304 UP000008792 UP000008711 UP000192221 UP000001070 UP000002282 UP000192223 UP000007266 UP000235965 UP000245037
UP000002320 UP000075883 UP000075920 UP000075900 UP000075886 UP000030765 UP000075880 UP000076408 UP000075903 UP000075884 UP000075902 UP000069272 UP000076407 UP000007062 UP000075840 UP000092462 UP000092461 UP000095301 UP000095300 UP000009192 UP000092553 UP000092445 UP000092443 UP000078200 UP000075882 UP000001819 UP000092460 UP000008744 UP000092444 UP000268350 UP000007798 UP000007801 UP000001292 UP000000803 UP000000304 UP000008792 UP000008711 UP000192221 UP000001070 UP000002282 UP000192223 UP000007266 UP000235965 UP000245037
Interpro
Gene 3D
ProteinModelPortal
H9JRH4
A0A194PFA0
A0A0N1IEK6
A0A212FC73
A0A2H1WI24
A0A2A4K4C3
+ More
A0A2A4K3L4 A0A2A4K3N6 A0A2A4K2L6 A0A1S4FBP6 Q178N6 A0A1S4G471 Q1DGT7 B0X888 A0A182M2G9 A0A182W5Q5 A0A182RHN8 A0A182R0S6 A0A084VQD2 A0A182J359 A0A182YCV2 A0A182UP81 A0A182N8I5 A0A182TUG8 A0A182FJ00 A0A182XJZ9 Q7QF99 A0A182IAX9 A0A1B0D8Y9 A0A1B0CHF1 T1PC13 A0A1I8ND94 A0A1I8Q1U1 B4L1M7 W8BDB3 A0A0M4EIM2 A0A1A9Z212 A0A1A9XZY6 A0A1A9VD56 A0A182KLC8 A0A0A1XRW6 A0A034V5G1 A0A034V863 Q29HR3 A0A1B0B403 B4GYG0 A0A0K8UQ93 A0A0K8UPG7 A0A1B0FAC1 A0A3B0JXW5 A0A0R3P4Z5 B4N1V3 B3MWG9 B4I9Y9 A0A0P9BV27 Q9W4W4 B4R3T9 B4MG75 B3NTW1 A0A1W4VF65 B4JL29 B4Q0X0 Q8MTX6 A0A1W4XAW8 A0A0R3P416 A0A139WEW0 A0A1Y1LLZ1 D6W9Y6 V5GSR3 O76862 D6WQJ0 A0A1W4XCK0 A0A1W4XCS1 A0A2J7QN57 A0A1W4WN84 A0A2P8Z4M8 A0A1E1WES9
A0A2A4K3L4 A0A2A4K3N6 A0A2A4K2L6 A0A1S4FBP6 Q178N6 A0A1S4G471 Q1DGT7 B0X888 A0A182M2G9 A0A182W5Q5 A0A182RHN8 A0A182R0S6 A0A084VQD2 A0A182J359 A0A182YCV2 A0A182UP81 A0A182N8I5 A0A182TUG8 A0A182FJ00 A0A182XJZ9 Q7QF99 A0A182IAX9 A0A1B0D8Y9 A0A1B0CHF1 T1PC13 A0A1I8ND94 A0A1I8Q1U1 B4L1M7 W8BDB3 A0A0M4EIM2 A0A1A9Z212 A0A1A9XZY6 A0A1A9VD56 A0A182KLC8 A0A0A1XRW6 A0A034V5G1 A0A034V863 Q29HR3 A0A1B0B403 B4GYG0 A0A0K8UQ93 A0A0K8UPG7 A0A1B0FAC1 A0A3B0JXW5 A0A0R3P4Z5 B4N1V3 B3MWG9 B4I9Y9 A0A0P9BV27 Q9W4W4 B4R3T9 B4MG75 B3NTW1 A0A1W4VF65 B4JL29 B4Q0X0 Q8MTX6 A0A1W4XAW8 A0A0R3P416 A0A139WEW0 A0A1Y1LLZ1 D6W9Y6 V5GSR3 O76862 D6WQJ0 A0A1W4XCK0 A0A1W4XCS1 A0A2J7QN57 A0A1W4WN84 A0A2P8Z4M8 A0A1E1WES9
PDB
4CA1
E-value=0.000568495,
Score=101
Ontologies
PATHWAY
GO
PANTHER
Topology
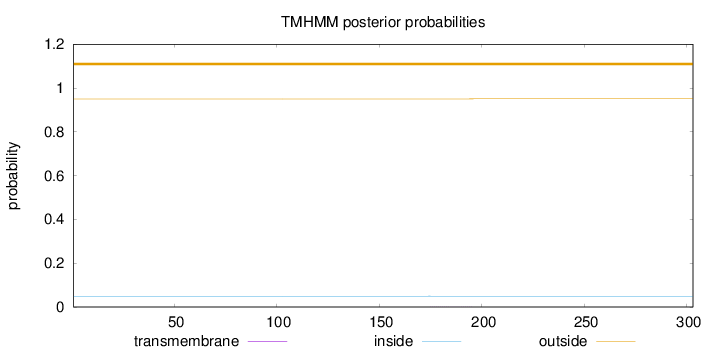
Length:
303
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01319
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04953
outside
1 - 303
Population Genetic Test Statistics
Pi
173.406131
Theta
165.551043
Tajima's D
-0.385769
CLR
1.787581
CSRT
0.270486475676216
Interpretation
Uncertain
