Gene
KWMTBOMO06736
Pre Gene Modal
BGIBMGA012136
Annotation
PREDICTED:_ADP-ribosylation_factor-like_protein_4C_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.421 Mitochondrial Reliability : 2.066
Sequence
CDS
ATGGGTGCGAATATGGGCAAAAACTCGAGCCTACTTGACGCGTTGCCTACCGCTCAGGGCACGCTCCACGTCGTCATGTTGGGCCTCGACTCAGCAGGGAAAACTACTGCACTATACAGACTCAAATTTGATCAATACTTAAACACCGTCCCCACTATTGGATTTAACTGCGAAAAGGTTAAAGGAACCATTGGAAAATCGAAGGGTGTGAACTTTCTAGTTTGGGATGTTGGCGGCCAGGAAAAACTAAGACCTCTTTGGAAATCGTACACACGTTGTACTGATGGTATAATTTTCGTATTAGACTCAGTCGACGTCGAACGAATGGAGGAAGCAAAAATGGAACTAATACGAACGGCAAAATCGCCAGATAATGCATGCGTGCCAATTCTAGTGTTAGCCAACAAACAGGATTTACCTGGGGCGAAAGAGCCTCGTGACCTCGAAAAGTTGCTTGGCCTGCATGAGCTGGTTTCTTCTCCACATGGCTCACGTGACGCACATGCGTGGCACGTTCAGCCAGCTTGTGCCATTACAGGGGAAGGCTTACACGAAGGTCTGGAAGTGCTTTACGAAATGATACTCAAGAGACGAAAACTGGCAAAGCTACAGAAAAAGCGCGTCAAATAA
Protein
MGANMGKNSSLLDALPTAQGTLHVVMLGLDSAGKTTALYRLKFDQYLNTVPTIGFNCEKVKGTIGKSKGVNFLVWDVGGQEKLRPLWKSYTRCTDGIIFVLDSVDVERMEEAKMELIRTAKSPDNACVPILVLANKQDLPGAKEPRDLEKLLGLHELVSSPHGSRDAHAWHVQPACAITGEGLHEGLEVLYEMILKRRKLAKLQKKRVK
Summary
Similarity
Belongs to the small GTPase superfamily. Arf family.
Uniprot
H9JRH6
A0A2A4K8Z2
A0A0L7L0A9
S4PRH6
A0A0N1PI23
A0A194PKR8
+ More
A0A1W4XIG6 A0A1B6IWZ7 A0A1B6GPE7 A0A067RDJ9 A0A1B6KTQ6 V5GH43 A0A212F5S8 A0A0P4VYY1 R4FN34 A0A224XPP7 A0A0V0GBK6 A0A023FB36 A0A069DQ04 M4VQI4 A0A1Y1JTK3 D6WGB7 A0A0A9WL49 A0A2P8Y1I4 A0A2R7VPK7 A0A0T6B5C9 E0VL35 A0A2R5LCV7 A0A1Z5LA82 A0A2H8U2X8 J9JTP7 A0A2S2P2Z6 A0A2S2QHM3 N6TH09 A0A164LDM0 A0A3S3P967 A0A1J1IIF4 A0A1L8DV90 A0A2P6KY69 A0A023GFV2 A0A1E1XQ60 A0A1E1WZD9 A0A087T1H7 A0A151IVY9 A0A026X021 F4WDM6 A0A195FUQ7 A0A158NXV9 A0A151WIC7 E2BZQ6 E2B0J7 E9G1Y4 A0A154P1D0 A0A087UA16 V9IF96 A0A088AM74 A0A131XEP9 A0A2P6JZ60 A0A224YH46 A0A131Z7T9 L7M0R8 A0A0P6BRF2 T1IKC8 K7IMS8 A0A0L7RF92 A0A1S3CW71 A0A2A3EAQ1 T1KBD2 A0A226EH43 A0A131Y0N1 B7PRK4 A0A1D2NKZ0 A0A151HZA9 T1GCK2 A0A034WAL1 A0A0N0U5H8 A0A0L0C3P2 A0A310S9B0 B4NHK1 T1KYC0 A0A1S3I5F9 A0A0P4ZFJ2 A0A1V9XCA1 B4MEX2 A0A0K8WL37 B4L7E5 A0A336L424 A0A0A1X2M7 B4JZQ5 B4H857 Q29CS8 A0A336MLS5 A0A0N8NZJ8 A0A1W4VKT0 R7VBW7 A0A0Q5SFE7 B3P9V4 H9XVM3 U5ERB8 A0A0J9US75
A0A1W4XIG6 A0A1B6IWZ7 A0A1B6GPE7 A0A067RDJ9 A0A1B6KTQ6 V5GH43 A0A212F5S8 A0A0P4VYY1 R4FN34 A0A224XPP7 A0A0V0GBK6 A0A023FB36 A0A069DQ04 M4VQI4 A0A1Y1JTK3 D6WGB7 A0A0A9WL49 A0A2P8Y1I4 A0A2R7VPK7 A0A0T6B5C9 E0VL35 A0A2R5LCV7 A0A1Z5LA82 A0A2H8U2X8 J9JTP7 A0A2S2P2Z6 A0A2S2QHM3 N6TH09 A0A164LDM0 A0A3S3P967 A0A1J1IIF4 A0A1L8DV90 A0A2P6KY69 A0A023GFV2 A0A1E1XQ60 A0A1E1WZD9 A0A087T1H7 A0A151IVY9 A0A026X021 F4WDM6 A0A195FUQ7 A0A158NXV9 A0A151WIC7 E2BZQ6 E2B0J7 E9G1Y4 A0A154P1D0 A0A087UA16 V9IF96 A0A088AM74 A0A131XEP9 A0A2P6JZ60 A0A224YH46 A0A131Z7T9 L7M0R8 A0A0P6BRF2 T1IKC8 K7IMS8 A0A0L7RF92 A0A1S3CW71 A0A2A3EAQ1 T1KBD2 A0A226EH43 A0A131Y0N1 B7PRK4 A0A1D2NKZ0 A0A151HZA9 T1GCK2 A0A034WAL1 A0A0N0U5H8 A0A0L0C3P2 A0A310S9B0 B4NHK1 T1KYC0 A0A1S3I5F9 A0A0P4ZFJ2 A0A1V9XCA1 B4MEX2 A0A0K8WL37 B4L7E5 A0A336L424 A0A0A1X2M7 B4JZQ5 B4H857 Q29CS8 A0A336MLS5 A0A0N8NZJ8 A0A1W4VKT0 R7VBW7 A0A0Q5SFE7 B3P9V4 H9XVM3 U5ERB8 A0A0J9US75
Pubmed
19121390
26227816
23622113
26354079
24845553
22118469
+ More
27129103 25474469 26334808 23497596 28004739 18362917 19820115 25401762 26823975 29403074 20566863 28528879 23537049 29209593 28503490 24508170 30249741 21719571 21347285 20798317 21292972 28049606 28797301 26830274 25576852 20075255 27289101 25348373 26108605 17994087 28327890 18057021 20479145 25830018 15632085 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249
27129103 25474469 26334808 23497596 28004739 18362917 19820115 25401762 26823975 29403074 20566863 28528879 23537049 29209593 28503490 24508170 30249741 21719571 21347285 20798317 21292972 28049606 28797301 26830274 25576852 20075255 27289101 25348373 26108605 17994087 28327890 18057021 20479145 25830018 15632085 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249
EMBL
BABH01016476
NWSH01000021
PCG80667.1
JTDY01003833
KOB68938.1
GAIX01000785
+ More
JAA91775.1 KQ460499 KPJ14281.1 KQ459606 KPI91695.1 GECU01016245 JAS91461.1 GECZ01032416 GECZ01030639 GECZ01026896 GECZ01025350 GECZ01024302 GECZ01024022 GECZ01018727 GECZ01016944 GECZ01009926 GECZ01008660 GECZ01006408 GECZ01005467 GECZ01003709 GECZ01003507 GECZ01001908 GECZ01000343 JAS37353.1 JAS39130.1 JAS42873.1 JAS44419.1 JAS45467.1 JAS45747.1 JAS51042.1 JAS52825.1 JAS59843.1 JAS61109.1 JAS63361.1 JAS64302.1 JAS66060.1 JAS66262.1 JAS67861.1 JAS69426.1 KK852533 KDR21842.1 GEBQ01025142 JAT14835.1 GALX01005137 JAB63329.1 AGBW02010142 OWR49092.1 GDKW01000773 JAI55822.1 ACPB03006731 ACPB03006732 GAHY01000723 JAA76787.1 GFTR01003372 JAW13054.1 GECL01000741 JAP05383.1 GBBI01000574 JAC18138.1 GBGD01002989 JAC85900.1 KC190027 AGI03912.1 GEZM01101487 GEZM01101486 JAV52632.1 KQ971329 EFA01144.1 GBHO01035090 GBRD01008073 GDHC01012678 JAG08514.1 JAG57748.1 JAQ05951.1 PYGN01001049 PSN38122.1 KK854004 PTY09058.1 LJIG01009728 KRT82500.1 DS235269 EEB14091.1 GGLE01003224 MBY07350.1 GFJQ02002755 JAW04215.1 GFXV01008133 MBW19938.1 ABLF02036596 GGMR01010647 MBY23266.1 GGMS01008035 MBY77238.1 APGK01038223 KB740954 KB632311 ENN77053.1 ERL92133.1 LRGB01003146 KZS04012.1 NCKU01000048 RWS17668.1 CVRI01000054 CRL00021.1 GFDF01003726 JAV10358.1 MWRG01003703 PRD31274.1 GBBM01002636 JAC32782.1 GFAA01001996 JAU01439.1 GFAC01006813 JAT92375.1 KK112970 KFM58966.1 KQ980882 KYN11960.1 KK107054 QOIP01000004 EZA61448.1 RLU24100.1 GL888089 EGI67687.1 KQ981261 KYN44156.1 ADTU01003628 KQ983089 KYQ47602.1 GL451657 EFN78818.1 GL444624 EFN60792.1 GL732529 EFX86726.1 KQ434796 KZC05735.1 KK118921 KFM74205.1 JR041010 AEY59342.1 GEFH01003092 JAP65489.1 MWRG01063234 PRD19361.1 GFPF01005840 MAA16986.1 GEDV01002326 JAP86231.1 GACK01007108 JAA57926.1 GDIP01011240 JAM92475.1 JH430488 KQ414608 KOC69386.1 KZ288313 PBC28372.1 CAEY01001946 LNIX01000003 OXA56983.1 GEFM01003771 JAP72025.1 ABJB010970353 DS773109 EEC09226.1 LJIJ01000013 ODN05948.1 KQ976709 KYM77056.1 CAQQ02031189 GAKP01008119 JAC50833.1 KQ435791 KOX74382.1 JRES01001020 KNC26094.1 KQ767621 OAD53368.1 CH964272 EDW83570.1 CAEY01000711 GDIP01214055 JAJ09347.1 MNPL01015284 OQR71147.1 CM000831 CH940665 ACY70541.1 EDW71073.1 GDHF01029965 GDHF01014519 GDHF01000527 JAI22349.1 JAI37795.1 JAI51787.1 CH933813 EDW10939.1 UFQS01001504 UFQT01001504 SSX11289.1 SSX30857.1 GBXI01009156 JAD05136.1 CH916380 EDV90921.1 CH479221 EDW34857.1 CH475413 EAL29334.2 UFQT01000750 SSX26958.1 CH902672 KPU74524.1 AMQN01004372 KB293367 ELU16057.1 CH954184 KQS21519.1 EDV45267.1 AE014135 AFH06768.1 GANO01003821 JAB56050.1 CM002916 KMZ07156.1
JAA91775.1 KQ460499 KPJ14281.1 KQ459606 KPI91695.1 GECU01016245 JAS91461.1 GECZ01032416 GECZ01030639 GECZ01026896 GECZ01025350 GECZ01024302 GECZ01024022 GECZ01018727 GECZ01016944 GECZ01009926 GECZ01008660 GECZ01006408 GECZ01005467 GECZ01003709 GECZ01003507 GECZ01001908 GECZ01000343 JAS37353.1 JAS39130.1 JAS42873.1 JAS44419.1 JAS45467.1 JAS45747.1 JAS51042.1 JAS52825.1 JAS59843.1 JAS61109.1 JAS63361.1 JAS64302.1 JAS66060.1 JAS66262.1 JAS67861.1 JAS69426.1 KK852533 KDR21842.1 GEBQ01025142 JAT14835.1 GALX01005137 JAB63329.1 AGBW02010142 OWR49092.1 GDKW01000773 JAI55822.1 ACPB03006731 ACPB03006732 GAHY01000723 JAA76787.1 GFTR01003372 JAW13054.1 GECL01000741 JAP05383.1 GBBI01000574 JAC18138.1 GBGD01002989 JAC85900.1 KC190027 AGI03912.1 GEZM01101487 GEZM01101486 JAV52632.1 KQ971329 EFA01144.1 GBHO01035090 GBRD01008073 GDHC01012678 JAG08514.1 JAG57748.1 JAQ05951.1 PYGN01001049 PSN38122.1 KK854004 PTY09058.1 LJIG01009728 KRT82500.1 DS235269 EEB14091.1 GGLE01003224 MBY07350.1 GFJQ02002755 JAW04215.1 GFXV01008133 MBW19938.1 ABLF02036596 GGMR01010647 MBY23266.1 GGMS01008035 MBY77238.1 APGK01038223 KB740954 KB632311 ENN77053.1 ERL92133.1 LRGB01003146 KZS04012.1 NCKU01000048 RWS17668.1 CVRI01000054 CRL00021.1 GFDF01003726 JAV10358.1 MWRG01003703 PRD31274.1 GBBM01002636 JAC32782.1 GFAA01001996 JAU01439.1 GFAC01006813 JAT92375.1 KK112970 KFM58966.1 KQ980882 KYN11960.1 KK107054 QOIP01000004 EZA61448.1 RLU24100.1 GL888089 EGI67687.1 KQ981261 KYN44156.1 ADTU01003628 KQ983089 KYQ47602.1 GL451657 EFN78818.1 GL444624 EFN60792.1 GL732529 EFX86726.1 KQ434796 KZC05735.1 KK118921 KFM74205.1 JR041010 AEY59342.1 GEFH01003092 JAP65489.1 MWRG01063234 PRD19361.1 GFPF01005840 MAA16986.1 GEDV01002326 JAP86231.1 GACK01007108 JAA57926.1 GDIP01011240 JAM92475.1 JH430488 KQ414608 KOC69386.1 KZ288313 PBC28372.1 CAEY01001946 LNIX01000003 OXA56983.1 GEFM01003771 JAP72025.1 ABJB010970353 DS773109 EEC09226.1 LJIJ01000013 ODN05948.1 KQ976709 KYM77056.1 CAQQ02031189 GAKP01008119 JAC50833.1 KQ435791 KOX74382.1 JRES01001020 KNC26094.1 KQ767621 OAD53368.1 CH964272 EDW83570.1 CAEY01000711 GDIP01214055 JAJ09347.1 MNPL01015284 OQR71147.1 CM000831 CH940665 ACY70541.1 EDW71073.1 GDHF01029965 GDHF01014519 GDHF01000527 JAI22349.1 JAI37795.1 JAI51787.1 CH933813 EDW10939.1 UFQS01001504 UFQT01001504 SSX11289.1 SSX30857.1 GBXI01009156 JAD05136.1 CH916380 EDV90921.1 CH479221 EDW34857.1 CH475413 EAL29334.2 UFQT01000750 SSX26958.1 CH902672 KPU74524.1 AMQN01004372 KB293367 ELU16057.1 CH954184 KQS21519.1 EDV45267.1 AE014135 AFH06768.1 GANO01003821 JAB56050.1 CM002916 KMZ07156.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000192223
+ More
UP000027135 UP000007151 UP000015103 UP000007266 UP000245037 UP000009046 UP000007819 UP000019118 UP000030742 UP000076858 UP000285301 UP000183832 UP000054359 UP000078492 UP000053097 UP000279307 UP000007755 UP000078541 UP000005205 UP000075809 UP000008237 UP000000311 UP000000305 UP000076502 UP000005203 UP000002358 UP000053825 UP000079169 UP000242457 UP000015104 UP000198287 UP000001555 UP000094527 UP000078540 UP000015102 UP000053105 UP000037069 UP000007798 UP000085678 UP000192247 UP000008792 UP000009192 UP000001070 UP000008744 UP000001819 UP000007801 UP000192221 UP000014760 UP000008711 UP000000803
UP000027135 UP000007151 UP000015103 UP000007266 UP000245037 UP000009046 UP000007819 UP000019118 UP000030742 UP000076858 UP000285301 UP000183832 UP000054359 UP000078492 UP000053097 UP000279307 UP000007755 UP000078541 UP000005205 UP000075809 UP000008237 UP000000311 UP000000305 UP000076502 UP000005203 UP000002358 UP000053825 UP000079169 UP000242457 UP000015104 UP000198287 UP000001555 UP000094527 UP000078540 UP000015102 UP000053105 UP000037069 UP000007798 UP000085678 UP000192247 UP000008792 UP000009192 UP000001070 UP000008744 UP000001819 UP000007801 UP000192221 UP000014760 UP000008711 UP000000803
PRIDE
Interpro
ProteinModelPortal
H9JRH6
A0A2A4K8Z2
A0A0L7L0A9
S4PRH6
A0A0N1PI23
A0A194PKR8
+ More
A0A1W4XIG6 A0A1B6IWZ7 A0A1B6GPE7 A0A067RDJ9 A0A1B6KTQ6 V5GH43 A0A212F5S8 A0A0P4VYY1 R4FN34 A0A224XPP7 A0A0V0GBK6 A0A023FB36 A0A069DQ04 M4VQI4 A0A1Y1JTK3 D6WGB7 A0A0A9WL49 A0A2P8Y1I4 A0A2R7VPK7 A0A0T6B5C9 E0VL35 A0A2R5LCV7 A0A1Z5LA82 A0A2H8U2X8 J9JTP7 A0A2S2P2Z6 A0A2S2QHM3 N6TH09 A0A164LDM0 A0A3S3P967 A0A1J1IIF4 A0A1L8DV90 A0A2P6KY69 A0A023GFV2 A0A1E1XQ60 A0A1E1WZD9 A0A087T1H7 A0A151IVY9 A0A026X021 F4WDM6 A0A195FUQ7 A0A158NXV9 A0A151WIC7 E2BZQ6 E2B0J7 E9G1Y4 A0A154P1D0 A0A087UA16 V9IF96 A0A088AM74 A0A131XEP9 A0A2P6JZ60 A0A224YH46 A0A131Z7T9 L7M0R8 A0A0P6BRF2 T1IKC8 K7IMS8 A0A0L7RF92 A0A1S3CW71 A0A2A3EAQ1 T1KBD2 A0A226EH43 A0A131Y0N1 B7PRK4 A0A1D2NKZ0 A0A151HZA9 T1GCK2 A0A034WAL1 A0A0N0U5H8 A0A0L0C3P2 A0A310S9B0 B4NHK1 T1KYC0 A0A1S3I5F9 A0A0P4ZFJ2 A0A1V9XCA1 B4MEX2 A0A0K8WL37 B4L7E5 A0A336L424 A0A0A1X2M7 B4JZQ5 B4H857 Q29CS8 A0A336MLS5 A0A0N8NZJ8 A0A1W4VKT0 R7VBW7 A0A0Q5SFE7 B3P9V4 H9XVM3 U5ERB8 A0A0J9US75
A0A1W4XIG6 A0A1B6IWZ7 A0A1B6GPE7 A0A067RDJ9 A0A1B6KTQ6 V5GH43 A0A212F5S8 A0A0P4VYY1 R4FN34 A0A224XPP7 A0A0V0GBK6 A0A023FB36 A0A069DQ04 M4VQI4 A0A1Y1JTK3 D6WGB7 A0A0A9WL49 A0A2P8Y1I4 A0A2R7VPK7 A0A0T6B5C9 E0VL35 A0A2R5LCV7 A0A1Z5LA82 A0A2H8U2X8 J9JTP7 A0A2S2P2Z6 A0A2S2QHM3 N6TH09 A0A164LDM0 A0A3S3P967 A0A1J1IIF4 A0A1L8DV90 A0A2P6KY69 A0A023GFV2 A0A1E1XQ60 A0A1E1WZD9 A0A087T1H7 A0A151IVY9 A0A026X021 F4WDM6 A0A195FUQ7 A0A158NXV9 A0A151WIC7 E2BZQ6 E2B0J7 E9G1Y4 A0A154P1D0 A0A087UA16 V9IF96 A0A088AM74 A0A131XEP9 A0A2P6JZ60 A0A224YH46 A0A131Z7T9 L7M0R8 A0A0P6BRF2 T1IKC8 K7IMS8 A0A0L7RF92 A0A1S3CW71 A0A2A3EAQ1 T1KBD2 A0A226EH43 A0A131Y0N1 B7PRK4 A0A1D2NKZ0 A0A151HZA9 T1GCK2 A0A034WAL1 A0A0N0U5H8 A0A0L0C3P2 A0A310S9B0 B4NHK1 T1KYC0 A0A1S3I5F9 A0A0P4ZFJ2 A0A1V9XCA1 B4MEX2 A0A0K8WL37 B4L7E5 A0A336L424 A0A0A1X2M7 B4JZQ5 B4H857 Q29CS8 A0A336MLS5 A0A0N8NZJ8 A0A1W4VKT0 R7VBW7 A0A0Q5SFE7 B3P9V4 H9XVM3 U5ERB8 A0A0J9US75
PDB
4FME
E-value=5.63108e-40,
Score=409
Ontologies
GO
Topology
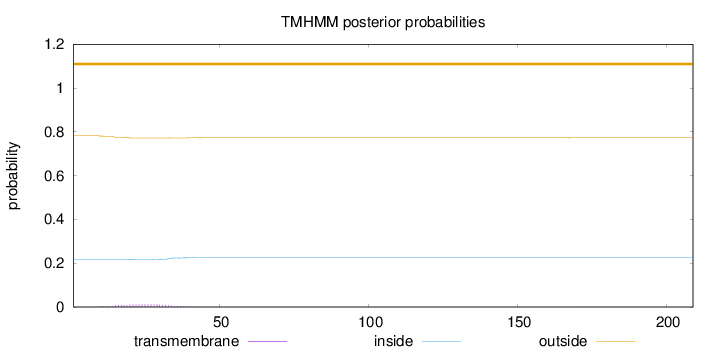
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24461
Exp number, first 60 AAs:
0.2403
Total prob of N-in:
0.21711
outside
1 - 209
Population Genetic Test Statistics
Pi
150.713235
Theta
156.684438
Tajima's D
-0.819362
CLR
0.115475
CSRT
0.171291435428229
Interpretation
Uncertain
