Pre Gene Modal
BGIBMGA012013
Annotation
PREDICTED:_ATP-dependent_RNA_helicase_p62-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.327
Sequence
CDS
ATGGCTTTAAATGTGCTCACAAGGTTATTAAAACGAAAGGAGTTGTCCAAATTATTACCTTACAAACAACAAATAGTAGAAACAAATGTATTTCACACTCACATTATTCCCAAGGTTCAATATTCTATATTGCACAATCAAATATTAAAAGGTACAAAACTACGAGAGCTACAAATCTTAAGATTTTATTCTGTAAAAACTGAGCAGAAAAAGGAAGATGATGATTACAGAAGTGATAATAGCATTACTGTTATTGGTCAAGATATACCCAGTCCTTTTGTTGACATAGAAGCCAGTGGATTTCCTGAATATATAAAAACATTTCTGAAGATGCAAGGTATCACAAAACCAACTGTGATACAGTCACAGGGATGGCCAATAGCTTTGAGTGGTAAAAACTTTGTGGGTATAGCACAAACAGGTACCGGAAAAACATTGGCTTTTCTTTTACCAGCTGTTGTTCATATTAAAGAAAATAAAGCAAAAAAGGGGCGAGGACCAATTGTTCTGGTTCTTGCCCCAACTAGGGAATTGGCAAGACAAATAGAGCAGGTGGCACAGGATTTTAATGAGCAATTAGGTATACGCTGTGCTTGTATTTACGGTGGAGCAAATAGAAGTAAACAAGCAGCACAGTTACAAGCTGGAGTTGACATAGTAATTGCAACTCCAGGAAGACTGAATGATTTTCTTATTACTAAGACAACAACATTAAGCAGGTGTACATATGTGGTGCTAGATGAAGCGGACCGAATGCTGGATATGGGTTTTGAACCTCAGATACGTCAAGCTTTAGAAGGAGTACCACTAGAGAGGCAAATCCTGATGTTCTCAGCAACATGGCCAAAGGAAGTCCGCACTCTTGCAAAAGATTATTTAGGTGAATTTGTCCAGGTCAATGTTGGTTCTACTGAGTTGTCTGCCAACCGCAACATACAACAAATTGTCCATGTATGTGAACAAGACGAAAAAGCCGAGAAGTTTACAGAGATAATGCATAAAATATCTGGTTTAGGTTTTGGTAAAACATTAGTATTTACAAACACTAAGAAATCTGTGGACTATCTTGAGAGAGTGCTTCGGGGAAATGGATGGCCAGCTCTAGGTATTCATGGGGATAGAACACAACTCCAGAGAGATATGATCATCAACAAATTTAAAACTGGAAAGACTAATATACTAGTAGCTACAGATGTTGCAGCTAGAGGTCTTGATGTTGATGGAGTGACACACGTAGTAAACTATGACTATCCCAATACATCTGAAGATTATATTCACAGAATAGGTAGAACAGGGCGTCAAGATAACAAGGGCATATCTCATTCTATACTCACTGAAGAAAATGCAAGACAGGCAAAGGATTTAATAGAAGTTTTGAAAGAAGCTAAACAGGATATCCCCAAAGAATTGTATGACTTGGCTCGCTCGTTTGACATCACGAAATCCACTGAAAAATTTTACAAAAAGAAAAGCTTCCAGAAGCCGTACAACTTTAGTAATAATGCGAGTAAAAAATGGAACAGATTTAGAAATGATAGATATTACTAA
Protein
MALNVLTRLLKRKELSKLLPYKQQIVETNVFHTHIIPKVQYSILHNQILKGTKLRELQILRFYSVKTEQKKEDDDYRSDNSITVIGQDIPSPFVDIEASGFPEYIKTFLKMQGITKPTVIQSQGWPIALSGKNFVGIAQTGTGKTLAFLLPAVVHIKENKAKKGRGPIVLVLAPTRELARQIEQVAQDFNEQLGIRCACIYGGANRSKQAAQLQAGVDIVIATPGRLNDFLITKTTTLSRCTYVVLDEADRMLDMGFEPQIRQALEGVPLERQILMFSATWPKEVRTLAKDYLGEFVQVNVGSTELSANRNIQQIVHVCEQDEKAEKFTEIMHKISGLGFGKTLVFTNTKKSVDYLERVLRGNGWPALGIHGDRTQLQRDMIINKFKTGKTNILVATDVAARGLDVDGVTHVVNYDYPNTSEDYIHRIGRTGRQDNKGISHSILTEENARQAKDLIEVLKEAKQDIPKELYDLARSFDITKSTEKFYKKKSFQKPYNFSNNASKKWNRFRNDRYY
Summary
Similarity
Belongs to the DEAD box helicase family.
Uniprot
H9JR53
A0A2A4K9Q0
A0A3S2PJ80
A0A194PKR3
A0A2H1WQD8
A0A0N1IPH2
+ More
A0A212F5T7 S4PMV6 A0A0L7L0Y4 A0A1B6BXY9 A0A1B6CV16 A0A0L0BWK4 A0A1B6C9I1 S4PFR6 A0A2H1WYV6 A0A1W4XIA1 A0A3G1T191 A0A139W8D1 A0A1W4WWH6 Q1HQ28 B2DBJ5 A0A194PG60 A0A139W8C8 V9IA90 A0A1W4XHA4 V9I9R9 A0A0L7QNQ4 A0A2A3E1Y1 D3TL05 A0A0N1IHL4 Q1HQ29 B4N2Y4 A0A1A9WBH0 A0A0P5XM62 A0A1B0FEH4 A0A0L7KP20 A0A2A4K913 A0A0P5UPA7 B4K198 I4DR20 A0A0P5SAE0 A0A0P5ZJ69 A0A154P740 A0A0P4YA91 H9JQE0 X1WJ61 A0A158NKG2 B3M9G0 F4W8R3 A0A0P5QD99 A0A1Y1L7K8 B4LF26 E9GQY5 A0A0P5ZRG9 A0A0N8P154 A0A0K8VWN6 A0A1Y1L2P5 A0A3L8D963 A0A195EHE1 A0A2J7QB95 A0A154P170 A0A310SD04 A0A0A9W0F9 A0A2J7QB62 A0A087ZXK7 A0A158P1F8 A0A3B0K2M7 A0A026X2Y0 A0A0P5ED32 A0A0A1WXE6 A0A0P5G919 A0A224YI85 A0A0N8A3A5 A0A0P5YXT2 B5DRG4 B4KZU0 A0A1W4V8W9 A0A0L7R8Q6 A0A1W4UWK9 A0A195D124 A0A162DGK2 A0A067R366 A0A0P4WSG1 A0A1B0GM31 A0A1D1XEY3 E0VL41 A0A0L7R2V8 A0A2A3ET06 V9ILQ7 A0A0P5HZP4 A0A088AS59 A0A0P6B602 A0A2J7QB73 A0A131Z1J7 B4PJT1 A0A0R1DWB6 A0A195F313 A0A034VBD0 B3NFL0
A0A212F5T7 S4PMV6 A0A0L7L0Y4 A0A1B6BXY9 A0A1B6CV16 A0A0L0BWK4 A0A1B6C9I1 S4PFR6 A0A2H1WYV6 A0A1W4XIA1 A0A3G1T191 A0A139W8D1 A0A1W4WWH6 Q1HQ28 B2DBJ5 A0A194PG60 A0A139W8C8 V9IA90 A0A1W4XHA4 V9I9R9 A0A0L7QNQ4 A0A2A3E1Y1 D3TL05 A0A0N1IHL4 Q1HQ29 B4N2Y4 A0A1A9WBH0 A0A0P5XM62 A0A1B0FEH4 A0A0L7KP20 A0A2A4K913 A0A0P5UPA7 B4K198 I4DR20 A0A0P5SAE0 A0A0P5ZJ69 A0A154P740 A0A0P4YA91 H9JQE0 X1WJ61 A0A158NKG2 B3M9G0 F4W8R3 A0A0P5QD99 A0A1Y1L7K8 B4LF26 E9GQY5 A0A0P5ZRG9 A0A0N8P154 A0A0K8VWN6 A0A1Y1L2P5 A0A3L8D963 A0A195EHE1 A0A2J7QB95 A0A154P170 A0A310SD04 A0A0A9W0F9 A0A2J7QB62 A0A087ZXK7 A0A158P1F8 A0A3B0K2M7 A0A026X2Y0 A0A0P5ED32 A0A0A1WXE6 A0A0P5G919 A0A224YI85 A0A0N8A3A5 A0A0P5YXT2 B5DRG4 B4KZU0 A0A1W4V8W9 A0A0L7R8Q6 A0A1W4UWK9 A0A195D124 A0A162DGK2 A0A067R366 A0A0P4WSG1 A0A1B0GM31 A0A1D1XEY3 E0VL41 A0A0L7R2V8 A0A2A3ET06 V9ILQ7 A0A0P5HZP4 A0A088AS59 A0A0P6B602 A0A2J7QB73 A0A131Z1J7 B4PJT1 A0A0R1DWB6 A0A195F313 A0A034VBD0 B3NFL0
Pubmed
EMBL
BABH01016476
NWSH01000021
PCG80696.1
RSAL01000016
RVE52988.1
KQ459606
+ More
KPI91690.1 ODYU01010215 SOQ55182.1 KQ460499 KPJ14286.1 AGBW02010142 OWR49088.1 GAIX01003350 JAA89210.1 JTDY01003833 KOB68939.1 GEDC01031146 JAS06152.1 GEDC01020087 JAS17211.1 JRES01001238 KNC24400.1 GEDC01027214 JAS10084.1 GAIX01003907 JAA88653.1 ODYU01012134 SOQ58265.1 MG846898 AXY94750.1 KQ973453 KXZ75521.1 DQ443224 ABF51313.1 AB264678 BAG30754.1 KPI91689.1 KXZ75522.1 JR036777 AEY57391.1 JR036778 AEY57392.1 KQ414851 KOC60199.1 KZ288518 PBC25101.1 EZ422095 ADD18300.1 KPJ14287.1 DQ443223 ABF51312.1 CH964062 EDW78723.2 GDIP01072458 JAM31257.1 CCAG010013880 JTDY01008184 KOB64694.1 PCG80697.1 GDIP01112144 JAL91570.1 CH916887 EDV90436.1 AK404897 BAM20360.1 GDIP01142344 JAL61370.1 GDIP01043664 JAM60051.1 KQ434829 KZC07755.1 GDIP01230427 JAI92974.1 BABH01011897 ABLF02031347 ABLF02031351 ABLF02031356 ABLF02049062 ADTU01018821 ADTU01018822 CH902618 EDV41173.1 GL887908 EGI69554.1 GDIQ01115740 JAL35986.1 GEZM01068283 JAV67047.1 CH940647 EDW69195.1 GL732558 EFX78251.1 GDIP01043665 JAM60050.1 KPU78972.1 GDHF01009026 JAI43288.1 GEZM01068282 JAV67048.1 QOIP01000011 RLU16672.1 KQ978881 KYN27690.1 NEVH01016301 PNF25839.1 KQ434796 KZC05689.1 KQ763581 OAD54947.1 GBHO01042703 GBHO01034983 GBRD01016935 GBRD01016932 JAG00901.1 JAG08621.1 JAG48892.1 PNF25838.1 ADTU01006523 OUUW01000012 SPP87573.1 KK107021 EZA62386.1 GDIP01148155 JAJ75247.1 GBXI01010755 JAD03537.1 GDIQ01266073 JAJ85651.1 GFPF01006231 MAA17377.1 GDIP01186697 JAJ36705.1 GDIP01051760 JAM51955.1 CH379070 EDY74133.1 CH933809 EDW17952.1 KQ414631 KOC67255.1 KQ976986 KYN06610.1 LRGB01001581 KZS11494.1 KK852729 KDR17555.1 GDIP01251427 JAI71974.1 AJVK01023899 AJVK01023900 AJVK01023901 GDJX01026979 JAT40957.1 DS235269 EEB14097.1 KQ414666 KOC65111.1 KZ288186 PBC34810.1 JR050357 AEY61249.1 GDIQ01226493 JAK25232.1 GDIP01019708 JAM84007.1 PNF25833.1 GEDV01004221 JAP84336.1 CM000159 EDW93680.1 KRK01301.1 KQ981852 KYN34965.1 GAKP01019541 GAKP01019540 JAC39412.1 CH954178 EDV50552.1
KPI91690.1 ODYU01010215 SOQ55182.1 KQ460499 KPJ14286.1 AGBW02010142 OWR49088.1 GAIX01003350 JAA89210.1 JTDY01003833 KOB68939.1 GEDC01031146 JAS06152.1 GEDC01020087 JAS17211.1 JRES01001238 KNC24400.1 GEDC01027214 JAS10084.1 GAIX01003907 JAA88653.1 ODYU01012134 SOQ58265.1 MG846898 AXY94750.1 KQ973453 KXZ75521.1 DQ443224 ABF51313.1 AB264678 BAG30754.1 KPI91689.1 KXZ75522.1 JR036777 AEY57391.1 JR036778 AEY57392.1 KQ414851 KOC60199.1 KZ288518 PBC25101.1 EZ422095 ADD18300.1 KPJ14287.1 DQ443223 ABF51312.1 CH964062 EDW78723.2 GDIP01072458 JAM31257.1 CCAG010013880 JTDY01008184 KOB64694.1 PCG80697.1 GDIP01112144 JAL91570.1 CH916887 EDV90436.1 AK404897 BAM20360.1 GDIP01142344 JAL61370.1 GDIP01043664 JAM60051.1 KQ434829 KZC07755.1 GDIP01230427 JAI92974.1 BABH01011897 ABLF02031347 ABLF02031351 ABLF02031356 ABLF02049062 ADTU01018821 ADTU01018822 CH902618 EDV41173.1 GL887908 EGI69554.1 GDIQ01115740 JAL35986.1 GEZM01068283 JAV67047.1 CH940647 EDW69195.1 GL732558 EFX78251.1 GDIP01043665 JAM60050.1 KPU78972.1 GDHF01009026 JAI43288.1 GEZM01068282 JAV67048.1 QOIP01000011 RLU16672.1 KQ978881 KYN27690.1 NEVH01016301 PNF25839.1 KQ434796 KZC05689.1 KQ763581 OAD54947.1 GBHO01042703 GBHO01034983 GBRD01016935 GBRD01016932 JAG00901.1 JAG08621.1 JAG48892.1 PNF25838.1 ADTU01006523 OUUW01000012 SPP87573.1 KK107021 EZA62386.1 GDIP01148155 JAJ75247.1 GBXI01010755 JAD03537.1 GDIQ01266073 JAJ85651.1 GFPF01006231 MAA17377.1 GDIP01186697 JAJ36705.1 GDIP01051760 JAM51955.1 CH379070 EDY74133.1 CH933809 EDW17952.1 KQ414631 KOC67255.1 KQ976986 KYN06610.1 LRGB01001581 KZS11494.1 KK852729 KDR17555.1 GDIP01251427 JAI71974.1 AJVK01023899 AJVK01023900 AJVK01023901 GDJX01026979 JAT40957.1 DS235269 EEB14097.1 KQ414666 KOC65111.1 KZ288186 PBC34810.1 JR050357 AEY61249.1 GDIQ01226493 JAK25232.1 GDIP01019708 JAM84007.1 PNF25833.1 GEDV01004221 JAP84336.1 CM000159 EDW93680.1 KRK01301.1 KQ981852 KYN34965.1 GAKP01019541 GAKP01019540 JAC39412.1 CH954178 EDV50552.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000037069 UP000192223 UP000007266 UP000053825 UP000242457 UP000007798 UP000091820 UP000092444 UP000001070 UP000076502 UP000007819 UP000005205 UP000007801 UP000007755 UP000008792 UP000000305 UP000279307 UP000078492 UP000235965 UP000005203 UP000268350 UP000053097 UP000001819 UP000009192 UP000192221 UP000078542 UP000076858 UP000027135 UP000092462 UP000009046 UP000002282 UP000078541 UP000008711
UP000037510 UP000037069 UP000192223 UP000007266 UP000053825 UP000242457 UP000007798 UP000091820 UP000092444 UP000001070 UP000076502 UP000007819 UP000005205 UP000007801 UP000007755 UP000008792 UP000000305 UP000279307 UP000078492 UP000235965 UP000005203 UP000268350 UP000053097 UP000001819 UP000009192 UP000192221 UP000078542 UP000076858 UP000027135 UP000092462 UP000009046 UP000002282 UP000078541 UP000008711
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9JR53
A0A2A4K9Q0
A0A3S2PJ80
A0A194PKR3
A0A2H1WQD8
A0A0N1IPH2
+ More
A0A212F5T7 S4PMV6 A0A0L7L0Y4 A0A1B6BXY9 A0A1B6CV16 A0A0L0BWK4 A0A1B6C9I1 S4PFR6 A0A2H1WYV6 A0A1W4XIA1 A0A3G1T191 A0A139W8D1 A0A1W4WWH6 Q1HQ28 B2DBJ5 A0A194PG60 A0A139W8C8 V9IA90 A0A1W4XHA4 V9I9R9 A0A0L7QNQ4 A0A2A3E1Y1 D3TL05 A0A0N1IHL4 Q1HQ29 B4N2Y4 A0A1A9WBH0 A0A0P5XM62 A0A1B0FEH4 A0A0L7KP20 A0A2A4K913 A0A0P5UPA7 B4K198 I4DR20 A0A0P5SAE0 A0A0P5ZJ69 A0A154P740 A0A0P4YA91 H9JQE0 X1WJ61 A0A158NKG2 B3M9G0 F4W8R3 A0A0P5QD99 A0A1Y1L7K8 B4LF26 E9GQY5 A0A0P5ZRG9 A0A0N8P154 A0A0K8VWN6 A0A1Y1L2P5 A0A3L8D963 A0A195EHE1 A0A2J7QB95 A0A154P170 A0A310SD04 A0A0A9W0F9 A0A2J7QB62 A0A087ZXK7 A0A158P1F8 A0A3B0K2M7 A0A026X2Y0 A0A0P5ED32 A0A0A1WXE6 A0A0P5G919 A0A224YI85 A0A0N8A3A5 A0A0P5YXT2 B5DRG4 B4KZU0 A0A1W4V8W9 A0A0L7R8Q6 A0A1W4UWK9 A0A195D124 A0A162DGK2 A0A067R366 A0A0P4WSG1 A0A1B0GM31 A0A1D1XEY3 E0VL41 A0A0L7R2V8 A0A2A3ET06 V9ILQ7 A0A0P5HZP4 A0A088AS59 A0A0P6B602 A0A2J7QB73 A0A131Z1J7 B4PJT1 A0A0R1DWB6 A0A195F313 A0A034VBD0 B3NFL0
A0A212F5T7 S4PMV6 A0A0L7L0Y4 A0A1B6BXY9 A0A1B6CV16 A0A0L0BWK4 A0A1B6C9I1 S4PFR6 A0A2H1WYV6 A0A1W4XIA1 A0A3G1T191 A0A139W8D1 A0A1W4WWH6 Q1HQ28 B2DBJ5 A0A194PG60 A0A139W8C8 V9IA90 A0A1W4XHA4 V9I9R9 A0A0L7QNQ4 A0A2A3E1Y1 D3TL05 A0A0N1IHL4 Q1HQ29 B4N2Y4 A0A1A9WBH0 A0A0P5XM62 A0A1B0FEH4 A0A0L7KP20 A0A2A4K913 A0A0P5UPA7 B4K198 I4DR20 A0A0P5SAE0 A0A0P5ZJ69 A0A154P740 A0A0P4YA91 H9JQE0 X1WJ61 A0A158NKG2 B3M9G0 F4W8R3 A0A0P5QD99 A0A1Y1L7K8 B4LF26 E9GQY5 A0A0P5ZRG9 A0A0N8P154 A0A0K8VWN6 A0A1Y1L2P5 A0A3L8D963 A0A195EHE1 A0A2J7QB95 A0A154P170 A0A310SD04 A0A0A9W0F9 A0A2J7QB62 A0A087ZXK7 A0A158P1F8 A0A3B0K2M7 A0A026X2Y0 A0A0P5ED32 A0A0A1WXE6 A0A0P5G919 A0A224YI85 A0A0N8A3A5 A0A0P5YXT2 B5DRG4 B4KZU0 A0A1W4V8W9 A0A0L7R8Q6 A0A1W4UWK9 A0A195D124 A0A162DGK2 A0A067R366 A0A0P4WSG1 A0A1B0GM31 A0A1D1XEY3 E0VL41 A0A0L7R2V8 A0A2A3ET06 V9ILQ7 A0A0P5HZP4 A0A088AS59 A0A0P6B602 A0A2J7QB73 A0A131Z1J7 B4PJT1 A0A0R1DWB6 A0A195F313 A0A034VBD0 B3NFL0
PDB
5E7M
E-value=2.32431e-80,
Score=762
Ontologies
GO
Topology
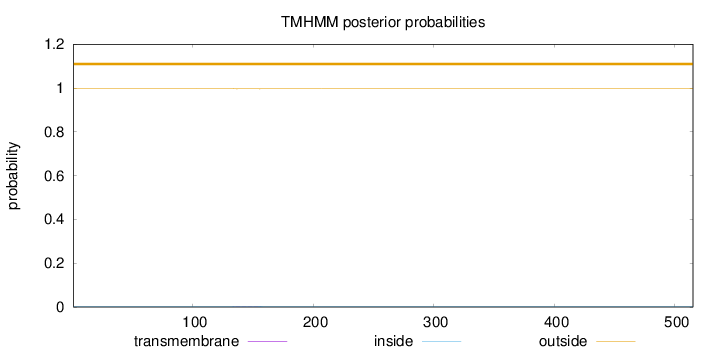
Length:
515
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0514600000000001
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00190
outside
1 - 515
Population Genetic Test Statistics
Pi
253.843979
Theta
219.614876
Tajima's D
0.111571
CLR
0.180573
CSRT
0.398530073496325
Interpretation
Uncertain
