Gene
KWMTBOMO06732 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011746
Annotation
DEAD_box_polypeptide_5_isoform_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.045 Nuclear Reliability : 1.347
Sequence
CDS
ATGGGTTACAAAGAACCGACGCCCATTCAAGCTCAAGGCTGGCCGATAGCTATGTCTGGAAAGAATTTAGTTGGCGTAGCCCAAACGGGTTCCGGCAAAACGTTGGCCTACATCTTGCCAGCCATTGTGCACATAAACAACCAACCGCCTATTCGGAGAGGTGATGGTCCGATTGCTTTGGTCTTGGCGCCTACCAGAGAGTTAGCACAACAAATTCAGCAAGTTGCTGCAGATTTTGGACACACATCTTATGTTCGTAACACGTGTGTGTTTGGTGGTGCTCCTAAAAGAGAGCAAGCCCGGGACTTGGAGAGGGGAGTAGAAATAGTCATTGCTACTCCAGGTAGATTAATTGATTTCTTGGAAAAGGGCACAACCAACTTACAGCGGTGCACATATTTAGTTCTTGATGAGGCTGATCGTATGTTGGATATGGGATTTGAACCACAAATCAGAAAAATCATTGAGCAAATACGCCCAGACAGACAGACTTTGATGTGGTCAGCTACTTGGCCCAAAGAAGTGAAGAAACTTGCTGAGGATTACTTGGGAGACTACATTCAGATCAATATAGGATCATTACAACTTTCCGCAAATCACAACATTCTTCAAATTGTAGATATTTGTCAAGAACATGAAAAAGAAAATAAATTAAATGTATTATTGCAAGAAATTGGACAAAGTCAAGAACCTGGTGCGAAAACAATAATTTTTGTTGAAACCAAGAGAAAAGCTGAGAACATATCAAGGAACATAAGGAGATATGGCTGGCCAGCTGTTTGCATGCATGGCGATAAAACTCAACAAGAAAGAGATGAAGTTCTGTATCAGTTCAAGGAAGGTCGTGCTAGTATTCTTGTAGCAACTGATGTTGCAGCTCGAGGGCTTGATGTTGATGGTATCAAATATGTAATAAATTTTGATTATCCAAATTCGTCGGAGGATTACATCCATCGTATTGGGAGAACTGGACGTTCAAAATCAAAAGGAACATCATATGCTTTCTTTACCCCTTCAAATTCCCGTCAAGCCAAAGATTTGGTATCTGTTCTACAAGAAGCTAATCAGATTATTAGCCCACAACTGCAGTCGATGGCCGACCGTTGCGGTGGTGGTGGCGGCGGGTGGAACAGGAACAGATTCGGCGGAGGTGGACGAGGTGGTGGCGGCTCTTTTAAACGCGGAAGCAATTTTGGCAAGAGTAGCCAAAGGGGGGGCAGTGGTTACAAACGATTTGACGACTATTAA
Protein
MGYKEPTPIQAQGWPIAMSGKNLVGVAQTGSGKTLAYILPAIVHINNQPPIRRGDGPIALVLAPTRELAQQIQQVAADFGHTSYVRNTCVFGGAPKREQARDLERGVEIVIATPGRLIDFLEKGTTNLQRCTYLVLDEADRMLDMGFEPQIRKIIEQIRPDRQTLMWSATWPKEVKKLAEDYLGDYIQINIGSLQLSANHNILQIVDICQEHEKENKLNVLLQEIGQSQEPGAKTIIFVETKRKAENISRNIRRYGWPAVCMHGDKTQQERDEVLYQFKEGRASILVATDVAARGLDVDGIKYVINFDYPNSSEDYIHRIGRTGRSKSKGTSYAFFTPSNSRQAKDLVSVLQEANQIISPQLQSMADRCGGGGGGWNRNRFGGGGRGGGGSFKRGSNFGKSSQRGGSGYKRFDDY
Summary
Similarity
Belongs to the DEAD box helicase family.
Uniprot
Q1HQ28
H9JQE0
Q1HQ29
A0A2H1WYV6
A0A194PG60
A0A2A4K913
+ More
B2DBJ5 I4DR20 A0A0N1IHL4 A0A3G1T191 A0A212F5T7 A0A0L7KP20 S4PFR6 A0A139W8D1 A0A0T6AUE2 A0A1Y1L7K8 A0A1Y1L2P5 A0A1W4XHA4 A0A1W4XIA1 V5GVC0 A0A139W8C8 A0A232FAC1 K7J4U7 A0A0T6AWR4 N6TXJ7 A0A154PM95 V5GQF2 U4UJK7 A0A067QTL2 A0A347ZJD9 A0A1W4WW60 A0A1W4WWH6 V5GUQ8 A0A067RCE2 A0A2J7QBB9 U4UMU0 A0A2J7PCX5 A0A067RF17 A0A2P8Z3M3 A0A2J7PCY0 K7J4U6 A0A232EJE7 A0A2A4K9X6 A0A0L7QNQ4 A0A2A4K9F6 A0A2J7QB73 A0A2A3E1Y1 V9I9R9 V9IA90 A0A0L7R2V8 A0A0N1IU74 A0A154P740 K7J871 A0A088AS59 A0A162DGK2 E9GQY5 A0A2A3ET06 V9ILQ7 E0VL41 A0A0P5ED32 E9J4I7 A0A0P5UPA7 A0A0P5ZJ69 A0A0P5XM62 A0A0P5SAE0 A0A0N8CUX9 A0A1B6DGM6 D6WGP2 A0A0J7NQS8 A0A087TNM1 A0A0P5ZRG9 W8BSP6 A0A0P5YXT2 A0A0P6B602 A0A0P5ZHP3 A0A0P5NA51 A0A0N7ZJJ9 A0A0N8EDI9 A0A1B6BXY9 A0A034WG23 A0A034WEI6 A0A0P5QD99 A0A0P5HZP4 A0A0N8A3A5 A0A026X1R8 A0A151IE74 E2B2Z3 A0A158NMB3 A0A151X1Y1 A0A151JLE1 F4WK10 A0A2A3EFJ0 A0A151JWH7 A0A158P1F8 A0A0P5VZ52 A0A0P4YA91 A0A1B6CV16 A0A3L8DU62 E9IJL8 A0A195F313
B2DBJ5 I4DR20 A0A0N1IHL4 A0A3G1T191 A0A212F5T7 A0A0L7KP20 S4PFR6 A0A139W8D1 A0A0T6AUE2 A0A1Y1L7K8 A0A1Y1L2P5 A0A1W4XHA4 A0A1W4XIA1 V5GVC0 A0A139W8C8 A0A232FAC1 K7J4U7 A0A0T6AWR4 N6TXJ7 A0A154PM95 V5GQF2 U4UJK7 A0A067QTL2 A0A347ZJD9 A0A1W4WW60 A0A1W4WWH6 V5GUQ8 A0A067RCE2 A0A2J7QBB9 U4UMU0 A0A2J7PCX5 A0A067RF17 A0A2P8Z3M3 A0A2J7PCY0 K7J4U6 A0A232EJE7 A0A2A4K9X6 A0A0L7QNQ4 A0A2A4K9F6 A0A2J7QB73 A0A2A3E1Y1 V9I9R9 V9IA90 A0A0L7R2V8 A0A0N1IU74 A0A154P740 K7J871 A0A088AS59 A0A162DGK2 E9GQY5 A0A2A3ET06 V9ILQ7 E0VL41 A0A0P5ED32 E9J4I7 A0A0P5UPA7 A0A0P5ZJ69 A0A0P5XM62 A0A0P5SAE0 A0A0N8CUX9 A0A1B6DGM6 D6WGP2 A0A0J7NQS8 A0A087TNM1 A0A0P5ZRG9 W8BSP6 A0A0P5YXT2 A0A0P6B602 A0A0P5ZHP3 A0A0P5NA51 A0A0N7ZJJ9 A0A0N8EDI9 A0A1B6BXY9 A0A034WG23 A0A034WEI6 A0A0P5QD99 A0A0P5HZP4 A0A0N8A3A5 A0A026X1R8 A0A151IE74 E2B2Z3 A0A158NMB3 A0A151X1Y1 A0A151JLE1 F4WK10 A0A2A3EFJ0 A0A151JWH7 A0A158P1F8 A0A0P5VZ52 A0A0P4YA91 A0A1B6CV16 A0A3L8DU62 E9IJL8 A0A195F313
Pubmed
EMBL
DQ443224
ABF51313.1
BABH01011897
DQ443223
ABF51312.1
ODYU01012134
+ More
SOQ58265.1 KQ459606 KPI91689.1 NWSH01000021 PCG80697.1 AB264678 BAG30754.1 AK404897 BAM20360.1 KQ460499 KPJ14287.1 MG846898 AXY94750.1 AGBW02010142 OWR49088.1 JTDY01008184 KOB64694.1 GAIX01003907 JAA88653.1 KQ973453 KXZ75521.1 LJIG01022795 KRT78682.1 GEZM01068283 JAV67047.1 GEZM01068282 JAV67048.1 GALX01002844 JAB65622.1 KXZ75522.1 NNAY01000614 OXU27383.1 AAZX01007814 LJIG01022617 KRT79603.1 APGK01057722 KB741280 KB632170 ENN71022.1 ERL89521.1 KQ434978 KZC13005.1 GALX01004619 JAB63847.1 KB632352 ERL93297.1 KK853035 KDR12221.1 FX985707 BBA84445.1 GALX01003064 JAB65402.1 KK852729 KDR17554.1 NEVH01016301 PNF25859.1 KB632390 ERL94437.1 NEVH01026401 PNF14191.1 KDR17563.1 PYGN01000211 PSN51101.1 PNF14190.1 NNAY01004047 OXU18442.1 PCG80698.1 KQ414851 KOC60199.1 PCG80699.1 PNF25833.1 KZ288518 PBC25101.1 JR036778 AEY57392.1 JR036777 AEY57391.1 KQ414666 KOC65111.1 KQ435693 KOX81002.1 KQ434829 KZC07755.1 LRGB01001581 KZS11494.1 GL732558 EFX78251.1 KZ288186 PBC34810.1 JR050357 AEY61249.1 DS235269 EEB14097.1 GDIP01148155 JAJ75247.1 GL768105 EFZ12253.1 GDIP01112144 JAL91570.1 GDIP01043664 JAM60051.1 GDIP01072458 JAM31257.1 GDIP01142344 JAL61370.1 GDIP01096415 JAM07300.1 GEDC01012518 JAS24780.1 KQ971329 EFA01112.2 LBMM01002449 KMQ94815.1 KK116065 KFM66710.1 GDIP01043665 JAM60050.1 GAMC01014331 JAB92224.1 GDIP01051760 JAM51955.1 GDIP01019708 JAM84007.1 GDIP01043396 JAM60319.1 GDIQ01145033 JAL06693.1 GDIP01239145 JAI84256.1 GDIQ01037356 JAN57381.1 GEDC01031146 JAS06152.1 GAKP01006259 JAC52693.1 GAKP01006260 JAC52692.1 GDIQ01115740 JAL35986.1 GDIQ01226493 JAK25232.1 GDIP01186697 JAJ36705.1 KK107024 EZA62250.1 KQ977890 KYM98981.1 GL445250 EFN89962.1 ADTU01020334 ADTU01020335 ADTU01020336 ADTU01020337 KQ982584 KYQ54410.1 KQ979011 KYN26621.1 GL888190 EGI65460.1 KZ288268 PBC30052.1 KQ981652 KYN38639.1 ADTU01006523 GDIP01094044 JAM09671.1 GDIP01230427 JAI92974.1 GEDC01020087 JAS17211.1 QOIP01000004 RLU23439.1 GL763868 EFZ19232.1 KQ981852 KYN34965.1
SOQ58265.1 KQ459606 KPI91689.1 NWSH01000021 PCG80697.1 AB264678 BAG30754.1 AK404897 BAM20360.1 KQ460499 KPJ14287.1 MG846898 AXY94750.1 AGBW02010142 OWR49088.1 JTDY01008184 KOB64694.1 GAIX01003907 JAA88653.1 KQ973453 KXZ75521.1 LJIG01022795 KRT78682.1 GEZM01068283 JAV67047.1 GEZM01068282 JAV67048.1 GALX01002844 JAB65622.1 KXZ75522.1 NNAY01000614 OXU27383.1 AAZX01007814 LJIG01022617 KRT79603.1 APGK01057722 KB741280 KB632170 ENN71022.1 ERL89521.1 KQ434978 KZC13005.1 GALX01004619 JAB63847.1 KB632352 ERL93297.1 KK853035 KDR12221.1 FX985707 BBA84445.1 GALX01003064 JAB65402.1 KK852729 KDR17554.1 NEVH01016301 PNF25859.1 KB632390 ERL94437.1 NEVH01026401 PNF14191.1 KDR17563.1 PYGN01000211 PSN51101.1 PNF14190.1 NNAY01004047 OXU18442.1 PCG80698.1 KQ414851 KOC60199.1 PCG80699.1 PNF25833.1 KZ288518 PBC25101.1 JR036778 AEY57392.1 JR036777 AEY57391.1 KQ414666 KOC65111.1 KQ435693 KOX81002.1 KQ434829 KZC07755.1 LRGB01001581 KZS11494.1 GL732558 EFX78251.1 KZ288186 PBC34810.1 JR050357 AEY61249.1 DS235269 EEB14097.1 GDIP01148155 JAJ75247.1 GL768105 EFZ12253.1 GDIP01112144 JAL91570.1 GDIP01043664 JAM60051.1 GDIP01072458 JAM31257.1 GDIP01142344 JAL61370.1 GDIP01096415 JAM07300.1 GEDC01012518 JAS24780.1 KQ971329 EFA01112.2 LBMM01002449 KMQ94815.1 KK116065 KFM66710.1 GDIP01043665 JAM60050.1 GAMC01014331 JAB92224.1 GDIP01051760 JAM51955.1 GDIP01019708 JAM84007.1 GDIP01043396 JAM60319.1 GDIQ01145033 JAL06693.1 GDIP01239145 JAI84256.1 GDIQ01037356 JAN57381.1 GEDC01031146 JAS06152.1 GAKP01006259 JAC52693.1 GAKP01006260 JAC52692.1 GDIQ01115740 JAL35986.1 GDIQ01226493 JAK25232.1 GDIP01186697 JAJ36705.1 KK107024 EZA62250.1 KQ977890 KYM98981.1 GL445250 EFN89962.1 ADTU01020334 ADTU01020335 ADTU01020336 ADTU01020337 KQ982584 KYQ54410.1 KQ979011 KYN26621.1 GL888190 EGI65460.1 KZ288268 PBC30052.1 KQ981652 KYN38639.1 ADTU01006523 GDIP01094044 JAM09671.1 GDIP01230427 JAI92974.1 GEDC01020087 JAS17211.1 QOIP01000004 RLU23439.1 GL763868 EFZ19232.1 KQ981852 KYN34965.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000037510
+ More
UP000007266 UP000192223 UP000215335 UP000002358 UP000019118 UP000030742 UP000076502 UP000027135 UP000235965 UP000245037 UP000053825 UP000242457 UP000053105 UP000005203 UP000076858 UP000000305 UP000009046 UP000036403 UP000054359 UP000053097 UP000078542 UP000008237 UP000005205 UP000075809 UP000078492 UP000007755 UP000078541 UP000279307
UP000007266 UP000192223 UP000215335 UP000002358 UP000019118 UP000030742 UP000076502 UP000027135 UP000235965 UP000245037 UP000053825 UP000242457 UP000053105 UP000005203 UP000076858 UP000000305 UP000009046 UP000036403 UP000054359 UP000053097 UP000078542 UP000008237 UP000005205 UP000075809 UP000078492 UP000007755 UP000078541 UP000279307
Interpro
IPR027417
P-loop_NTPase
+ More
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR025816 RrmJ-type_MeTrfase
IPR029063 SAM-dependent_MTases
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR030376 Cap_mRNA_MeTrfase_1
IPR000467 G_patch_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR025816 RrmJ-type_MeTrfase
IPR029063 SAM-dependent_MTases
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR030376 Cap_mRNA_MeTrfase_1
IPR000467 G_patch_dom
CDD
ProteinModelPortal
Q1HQ28
H9JQE0
Q1HQ29
A0A2H1WYV6
A0A194PG60
A0A2A4K913
+ More
B2DBJ5 I4DR20 A0A0N1IHL4 A0A3G1T191 A0A212F5T7 A0A0L7KP20 S4PFR6 A0A139W8D1 A0A0T6AUE2 A0A1Y1L7K8 A0A1Y1L2P5 A0A1W4XHA4 A0A1W4XIA1 V5GVC0 A0A139W8C8 A0A232FAC1 K7J4U7 A0A0T6AWR4 N6TXJ7 A0A154PM95 V5GQF2 U4UJK7 A0A067QTL2 A0A347ZJD9 A0A1W4WW60 A0A1W4WWH6 V5GUQ8 A0A067RCE2 A0A2J7QBB9 U4UMU0 A0A2J7PCX5 A0A067RF17 A0A2P8Z3M3 A0A2J7PCY0 K7J4U6 A0A232EJE7 A0A2A4K9X6 A0A0L7QNQ4 A0A2A4K9F6 A0A2J7QB73 A0A2A3E1Y1 V9I9R9 V9IA90 A0A0L7R2V8 A0A0N1IU74 A0A154P740 K7J871 A0A088AS59 A0A162DGK2 E9GQY5 A0A2A3ET06 V9ILQ7 E0VL41 A0A0P5ED32 E9J4I7 A0A0P5UPA7 A0A0P5ZJ69 A0A0P5XM62 A0A0P5SAE0 A0A0N8CUX9 A0A1B6DGM6 D6WGP2 A0A0J7NQS8 A0A087TNM1 A0A0P5ZRG9 W8BSP6 A0A0P5YXT2 A0A0P6B602 A0A0P5ZHP3 A0A0P5NA51 A0A0N7ZJJ9 A0A0N8EDI9 A0A1B6BXY9 A0A034WG23 A0A034WEI6 A0A0P5QD99 A0A0P5HZP4 A0A0N8A3A5 A0A026X1R8 A0A151IE74 E2B2Z3 A0A158NMB3 A0A151X1Y1 A0A151JLE1 F4WK10 A0A2A3EFJ0 A0A151JWH7 A0A158P1F8 A0A0P5VZ52 A0A0P4YA91 A0A1B6CV16 A0A3L8DU62 E9IJL8 A0A195F313
B2DBJ5 I4DR20 A0A0N1IHL4 A0A3G1T191 A0A212F5T7 A0A0L7KP20 S4PFR6 A0A139W8D1 A0A0T6AUE2 A0A1Y1L7K8 A0A1Y1L2P5 A0A1W4XHA4 A0A1W4XIA1 V5GVC0 A0A139W8C8 A0A232FAC1 K7J4U7 A0A0T6AWR4 N6TXJ7 A0A154PM95 V5GQF2 U4UJK7 A0A067QTL2 A0A347ZJD9 A0A1W4WW60 A0A1W4WWH6 V5GUQ8 A0A067RCE2 A0A2J7QBB9 U4UMU0 A0A2J7PCX5 A0A067RF17 A0A2P8Z3M3 A0A2J7PCY0 K7J4U6 A0A232EJE7 A0A2A4K9X6 A0A0L7QNQ4 A0A2A4K9F6 A0A2J7QB73 A0A2A3E1Y1 V9I9R9 V9IA90 A0A0L7R2V8 A0A0N1IU74 A0A154P740 K7J871 A0A088AS59 A0A162DGK2 E9GQY5 A0A2A3ET06 V9ILQ7 E0VL41 A0A0P5ED32 E9J4I7 A0A0P5UPA7 A0A0P5ZJ69 A0A0P5XM62 A0A0P5SAE0 A0A0N8CUX9 A0A1B6DGM6 D6WGP2 A0A0J7NQS8 A0A087TNM1 A0A0P5ZRG9 W8BSP6 A0A0P5YXT2 A0A0P6B602 A0A0P5ZHP3 A0A0P5NA51 A0A0N7ZJJ9 A0A0N8EDI9 A0A1B6BXY9 A0A034WG23 A0A034WEI6 A0A0P5QD99 A0A0P5HZP4 A0A0N8A3A5 A0A026X1R8 A0A151IE74 E2B2Z3 A0A158NMB3 A0A151X1Y1 A0A151JLE1 F4WK10 A0A2A3EFJ0 A0A151JWH7 A0A158P1F8 A0A0P5VZ52 A0A0P4YA91 A0A1B6CV16 A0A3L8DU62 E9IJL8 A0A195F313
PDB
2I4I
E-value=3.61457e-89,
Score=837
Ontologies
KEGG
GO
PANTHER
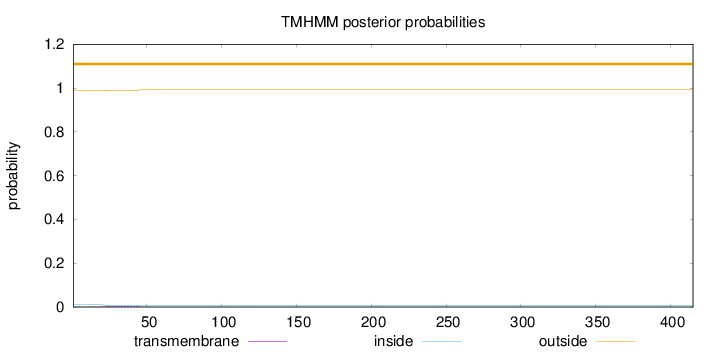
Topology
Length:
415
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11185
Exp number, first 60 AAs:
0.11185
Total prob of N-in:
0.01042
outside
1 - 415
Population Genetic Test Statistics
Pi
239.739
Theta
205.824879
Tajima's D
0.518704
CLR
3.8287
CSRT
0.522373881305935
Interpretation
Uncertain
