Pre Gene Modal
BGIBMGA011747
Annotation
PREDICTED:_histone_deacetylase_Rpd3_[Papilio_polytes]
Full name
Histone deacetylase
+ More
Histone deacetylase Rpd3
Histone deacetylase Rpd3
Location in the cell
Nuclear Reliability : 4.489
Sequence
CDS
ATGTCTATGCAACCGCACAGTAAGAAAAGAGTGTGTTACTATTATGACAGCGATATTGGAAACTACTATTACGGACAAGGGCATCCAATGAAACCTCACCGCATACGTATGACCCACAATTTACTCCTCAACTATGGACTCTATCGAAAAATGGAGATTTATAGGCCTCACAAAGCCACAGCTGATGAAATGACAAAATTCCATTCAGATGATTACATTAGATTCCTGCGTTCTATTCGACCTGACAATGTTTCTGAGTACAACAAACAAATGCAGAGATTTAATGTAGGTGAAGATTGTCCTGTTTTTGATGGCCTGTATGAATTTTGTCAGTTGTCTGCTGGGGGTTCAGTTGCAGCAGCTGTAAAACTGAATAAGCAGGCCTCAGAAATATGCATAAACTGGGGTGGTGGCCTCCATCATGCAAAGAAGTCTGAAGCGTCAGGTTTCTGTTATGTTAATGACATTGTTCTTGGCATATTGGAGTTGCTGAAATACCATCAAAGAGTACTATATATTGACATAGATGTGCATCATGGTGATGGTGTTGAAGAAGCTTTCTACACCACGGACAGAGTAATGACCGTTTCCTTTCATAAATATGGAGAATATTTTCCTGGAACAGGTGATTTGCGGGACATTGGTGCTGGTAAAGGCAAATACTATGCTGTAAATATACCCCTTCGTGATGGAATGGATGATGAGTCATATGAGTCTATCTTTGTTCCCATTATATCTAAAGTTATGGAGACATTCCAACCAAGTGCTGTAGTACTACAGTGTGGTGCTGATTCTCTGACAGGTGACAGATTGGGTTGTTTCAACTTGACTGTTCGAGGTCATGGACGCTGTGTAGAACTTGTCAAGAGGTTTGGACTTCCATTCTTACTAGTCGGCGGAGGCGGATATACCATACGTAATGTATCTCGCTGTTGGACTTACGAGACATCTGTTGCACTTGGTGTGGAAATAGCCAATGAACTACCATATAATGACTACTTTGAGTACTTTGGGCCCGATTTCAAACTACACATATCACCAAGTAACATGTCCAATCAAAACACACCCGAATATCTGGAAAAGATAAAAAATAGACTGTTCGAGAATCTCAGAATGCTCCCACATGCGCCAGGAGTACAAGTGCAAGCCATACCTGAAGATGCTGTCAATGATGAGTCAGATGATGAAGATAAAATAGACAAAGATGAGAGATTACCACAGAGTGATAAGGACAAGCGCATCGCTAACGACGGCGAGCTGTCGGACTCCGAGGACGAGGGCGACGGGCGACGCGACAACCGAGCCTACCGCGCGCCGCAGCGCAAGCGTCCGCGACTCGACAAGGAACCCTTGCAGATCAAGGACGATCTTAAACCTGATGACATGAAGGATGACGTTAAAAACGTTAGCAACGCGGAAGAATCAAAGAAGGACATGCCACCGAATCCCTGA
Protein
MSMQPHSKKRVCYYYDSDIGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKATADEMTKFHSDDYIRFLRSIRPDNVSEYNKQMQRFNVGEDCPVFDGLYEFCQLSAGGSVAAAVKLNKQASEICINWGGGLHHAKKSEASGFCYVNDIVLGILELLKYHQRVLYIDIDVHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNIPLRDGMDDESYESIFVPIISKVMETFQPSAVVLQCGADSLTGDRLGCFNLTVRGHGRCVELVKRFGLPFLLVGGGGYTIRNVSRCWTYETSVALGVEIANELPYNDYFEYFGPDFKLHISPSNMSNQNTPEYLEKIKNRLFENLRMLPHAPGVQVQAIPEDAVNDESDDEDKIDKDERLPQSDKDKRIANDGELSDSEDEGDGRRDNRAYRAPQRKRPRLDKEPLQIKDDLKPDDMKDDVKNVSNAEESKKDMPPNP
Summary
Description
Catalyzes the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4) (PubMed:11571273, PubMed:28245922, PubMed:12408863). Histone deacetylation may constitute a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events (PubMed:11571273, PubMed:8955276, PubMed:15545624, PubMed:15306652). For instance, deacetylation of histone H3 may be a prerequisite for the subsequent recruitment of the histone methyltransferase Su(var)3-9 to histones (PubMed:11571273). Involved in position-effect variegation (PEV) (PubMed:11571273). In the larval brain, part of a regulatory network including the transcriptional repressors klu, dpn and E(spl)mgamma-HLH which is required for type II neuroblast self-renewal and for maintaining erm in an inactive state in intermediate neural progenitors (INP) (PubMed:28245922).
Catalytic Activity
Hydrolysis of an N(6)-acetyl-lysine residue of a histone to yield a deacetylated histone.
Subunit
Interacts with Su(var)3-9. Component of a form of the Esc/E(z) complex present specifically during early embryogenesis which is composed of Caf1, esc, E(z), Su(z)12, Pcl and Rpd3. The Esc/E(z) complex may also associate with Pcl and Rpd3 during early embryogenesis. This complex is distinct from the PRC1 complex, which contains many other PcG proteins like Pc, Ph, Psc, Su(z)2. The 2 complexes however cooperate and interact together during the first 3 hours of development to establish PcG silencing. Interacts with the histone methyltransferase Su(var)3-9. Component of a complex that contains at least Rpd3, CoRest and Su(var)3-3/Hdm. Component of the DREAM complex at least composed of Myb, Caf1, mip40, mip120, mip130, E2f2, Dp, Rbf, Rbf2, lin-52, Rpd3 and l(3)mbt. Interacts with neuronal repressor Ttk88.
Similarity
Belongs to the histone deacetylase family. HD Type 1 subfamily.
Belongs to the histone deacetylase family. HD type 1 subfamily.
Belongs to the histone deacetylase family. HD type 1 subfamily.
Keywords
Chromatin regulator
Complete proteome
Developmental protein
Hydrolase
Nucleus
Phosphoprotein
Reference proteome
Repressor
Transcription
Transcription regulation
Feature
chain Histone deacetylase Rpd3
Uniprot
A0A2A4K903
A0A194PF89
A0A212F5S3
A0A0N1IGB9
A0A1Q3EXC7
B0WJ91
+ More
Q17CF0 A0A1Q3EXB1 A0A182GNN5 A0A023EUU4 B4MFY7 Q17CE9 A0A1Q3EXC6 U5EW58 B4IZ79 E3W281 E3W278 E3VX32 E3VX22 A0A0M4E8U0 B4MMY6 W8C8N5 A0A0J9RPK2 B4HU89 B3NC63 Q94517 B4QR08 A0A2M4AQV8 A0A1W4VAJ1 B4PIA1 A0A347ZJC4 A0A182FGH3 W5JF61 B3M9I7 A0A034WDK2 A0A0J7L4B5 A0A0A1WN83 B4KX99 A0A0K8VJ62 Q29EB4 A0A084WMI3 A0A182WBY2 A0A1Y1NG16 E3W282 S4NV65 A0A1I8M0Y6 A0A3B0JW31 A0A3B0J6G3 E2AL67 A0A182IKH6 A0A2J7QBD0 A0A336MS89 A0A182Y7V6 A0A1B0FJF6 A0A1A9Y199 A0A1A9UXP8 A0A1B0AZ17 A0A1A9ZWK4 A0A182V3V5 D6WG88 A0A182K1D6 A0A1L7N254 A0A182LKN9 A0A182TYI1 A0A182I495 A0A182X957 A0A182MWC0 A0A182RR67 A0A088AP92 E2B313 A0A195FTX4 A0A026WND6 A0A151I2N9 A0A158NKZ2 I1V501 A0A1I8PP67 Q7Q5D2 A0A151IM03 A0A151X3P9 J3JWV8 A0A151JAD5 F4WWT9 N6T0I9 A0A1L8DX14 A0A1A9W4R5 A0A1B6LWH4 B4HCY7 A0A1B6FBN9 A0A182QB58 A0A182NIY3 A0A232FI77 A0A3B0K2G2 A0A0R3P6D5 A0A1B6D3Z1 A0A0R3P695 K7J9J6 A0A182T387 A0A182NZT1 A0A2A3ESP9 A0A0A9YBV7
Q17CF0 A0A1Q3EXB1 A0A182GNN5 A0A023EUU4 B4MFY7 Q17CE9 A0A1Q3EXC6 U5EW58 B4IZ79 E3W281 E3W278 E3VX32 E3VX22 A0A0M4E8U0 B4MMY6 W8C8N5 A0A0J9RPK2 B4HU89 B3NC63 Q94517 B4QR08 A0A2M4AQV8 A0A1W4VAJ1 B4PIA1 A0A347ZJC4 A0A182FGH3 W5JF61 B3M9I7 A0A034WDK2 A0A0J7L4B5 A0A0A1WN83 B4KX99 A0A0K8VJ62 Q29EB4 A0A084WMI3 A0A182WBY2 A0A1Y1NG16 E3W282 S4NV65 A0A1I8M0Y6 A0A3B0JW31 A0A3B0J6G3 E2AL67 A0A182IKH6 A0A2J7QBD0 A0A336MS89 A0A182Y7V6 A0A1B0FJF6 A0A1A9Y199 A0A1A9UXP8 A0A1B0AZ17 A0A1A9ZWK4 A0A182V3V5 D6WG88 A0A182K1D6 A0A1L7N254 A0A182LKN9 A0A182TYI1 A0A182I495 A0A182X957 A0A182MWC0 A0A182RR67 A0A088AP92 E2B313 A0A195FTX4 A0A026WND6 A0A151I2N9 A0A158NKZ2 I1V501 A0A1I8PP67 Q7Q5D2 A0A151IM03 A0A151X3P9 J3JWV8 A0A151JAD5 F4WWT9 N6T0I9 A0A1L8DX14 A0A1A9W4R5 A0A1B6LWH4 B4HCY7 A0A1B6FBN9 A0A182QB58 A0A182NIY3 A0A232FI77 A0A3B0K2G2 A0A0R3P6D5 A0A1B6D3Z1 A0A0R3P695 K7J9J6 A0A182T387 A0A182NZT1 A0A2A3ESP9 A0A0A9YBV7
EC Number
3.5.1.98
Pubmed
26354079
22118469
17510324
26483478
24945155
17994087
+ More
24495485 22936249 8955276 10655219 10731132 12537572 12537569 11571273 12533794 12408863 12697833 15545624 15306652 18327897 28245922 17550304 20920257 23761445 25348373 25830018 15632085 24438588 28004739 23622113 25315136 20798317 25244985 18362917 19820115 27956627 20966253 24508170 30249741 21347285 12364791 22516182 23537049 21719571 28648823 20075255 25401762 26823975
24495485 22936249 8955276 10655219 10731132 12537572 12537569 11571273 12533794 12408863 12697833 15545624 15306652 18327897 28245922 17550304 20920257 23761445 25348373 25830018 15632085 24438588 28004739 23622113 25315136 20798317 25244985 18362917 19820115 27956627 20966253 24508170 30249741 21347285 12364791 22516182 23537049 21719571 28648823 20075255 25401762 26823975
EMBL
NWSH01000021
PCG80701.1
KQ459606
KPI91688.1
AGBW02010142
OWR49087.1
+ More
KQ460499 KPJ14288.1 GFDL01015083 JAV19962.1 DS231957 EDS29016.1 CH477309 EAT44001.1 GFDL01015100 JAV19945.1 JXUM01014757 JXUM01014758 JXUM01014759 JXUM01014760 KQ560412 KXJ82549.1 GAPW01001354 JAC12244.1 CH940669 EDW58248.1 EAT44002.1 GFDL01015081 JAV19964.1 GANO01003103 JAB56768.1 CH916366 EDV95601.1 HQ379193 ADO79638.1 HQ379190 ADO79635.1 HQ326584 ADO79632.1 HQ323752 HQ379189 HQ379191 HQ379192 ADO79633.1 ADO79634.1 ADO79636.1 ADO79637.1 CP012525 ALC43366.1 CH963847 EDW73542.1 GAMC01007366 GAMC01007365 JAB99189.1 CM002912 KMY97652.1 CH480817 EDW50510.1 CH954178 EDV50951.1 Y09258 AF086715 AF026949 AE014296 AY058487 CM000363 EDX09245.1 GGFK01009838 MBW43159.1 CM000159 EDW93443.1 FX985692 BBA84430.1 ADMH02001685 ETN61439.1 CH902618 EDV38993.1 GAKP01006198 JAC52754.1 LBMM01000796 KMQ97471.1 GBXI01014146 JAD00146.1 CH933809 EDW17557.1 GDHF01013412 JAI38902.1 CH379070 EAL30147.2 ATLV01024463 KE525352 KFB51427.1 GEZM01007639 JAV95026.1 HQ379194 ADO79639.1 GAIX01011651 JAA80909.1 OUUW01000002 SPP77576.1 SPP77577.1 GL440492 EFN65822.1 NEVH01016301 PNF25885.1 UFQT01001840 UFQT01003493 SSX31969.1 SSX35065.1 CCAG010004434 JXJN01006110 KQ971328 EEZ99767.1 LC096257 BAW19561.1 APCN01002287 AXCM01012496 GL445250 EFN89982.1 KQ981281 KYN43339.1 KK107152 QOIP01000007 EZA57151.1 RLU20012.1 KQ976546 KYM80955.1 ADTU01019057 JQ663527 AFI26262.1 AAAB01008960 EAA11382.2 KQ977085 KYN05918.1 KQ982557 KYQ55022.1 APGK01049592 APGK01049593 APGK01049594 BT127726 KB741156 KB632169 AEE62688.1 ENN73570.1 ERL89485.1 KQ979308 KYN21971.1 GL888413 EGI61327.1 ENN73569.1 GFDF01003084 JAV11000.1 GEBQ01011935 JAT28042.1 CH479537 EDW33672.1 GECZ01029352 GECZ01022436 GECZ01015271 JAS40417.1 JAS47333.1 JAS54498.1 AXCN02000463 NNAY01000158 OXU30451.1 SPP77578.1 KRT08680.1 GEDC01016932 GEDC01014994 JAS20366.1 JAS22304.1 KRT08681.1 AAZX01002696 KZ288193 PBC34071.1 GBHO01013975 GBRD01013839 GDHC01019818 JAG29629.1 JAG51987.1 JAP98810.1
KQ460499 KPJ14288.1 GFDL01015083 JAV19962.1 DS231957 EDS29016.1 CH477309 EAT44001.1 GFDL01015100 JAV19945.1 JXUM01014757 JXUM01014758 JXUM01014759 JXUM01014760 KQ560412 KXJ82549.1 GAPW01001354 JAC12244.1 CH940669 EDW58248.1 EAT44002.1 GFDL01015081 JAV19964.1 GANO01003103 JAB56768.1 CH916366 EDV95601.1 HQ379193 ADO79638.1 HQ379190 ADO79635.1 HQ326584 ADO79632.1 HQ323752 HQ379189 HQ379191 HQ379192 ADO79633.1 ADO79634.1 ADO79636.1 ADO79637.1 CP012525 ALC43366.1 CH963847 EDW73542.1 GAMC01007366 GAMC01007365 JAB99189.1 CM002912 KMY97652.1 CH480817 EDW50510.1 CH954178 EDV50951.1 Y09258 AF086715 AF026949 AE014296 AY058487 CM000363 EDX09245.1 GGFK01009838 MBW43159.1 CM000159 EDW93443.1 FX985692 BBA84430.1 ADMH02001685 ETN61439.1 CH902618 EDV38993.1 GAKP01006198 JAC52754.1 LBMM01000796 KMQ97471.1 GBXI01014146 JAD00146.1 CH933809 EDW17557.1 GDHF01013412 JAI38902.1 CH379070 EAL30147.2 ATLV01024463 KE525352 KFB51427.1 GEZM01007639 JAV95026.1 HQ379194 ADO79639.1 GAIX01011651 JAA80909.1 OUUW01000002 SPP77576.1 SPP77577.1 GL440492 EFN65822.1 NEVH01016301 PNF25885.1 UFQT01001840 UFQT01003493 SSX31969.1 SSX35065.1 CCAG010004434 JXJN01006110 KQ971328 EEZ99767.1 LC096257 BAW19561.1 APCN01002287 AXCM01012496 GL445250 EFN89982.1 KQ981281 KYN43339.1 KK107152 QOIP01000007 EZA57151.1 RLU20012.1 KQ976546 KYM80955.1 ADTU01019057 JQ663527 AFI26262.1 AAAB01008960 EAA11382.2 KQ977085 KYN05918.1 KQ982557 KYQ55022.1 APGK01049592 APGK01049593 APGK01049594 BT127726 KB741156 KB632169 AEE62688.1 ENN73570.1 ERL89485.1 KQ979308 KYN21971.1 GL888413 EGI61327.1 ENN73569.1 GFDF01003084 JAV11000.1 GEBQ01011935 JAT28042.1 CH479537 EDW33672.1 GECZ01029352 GECZ01022436 GECZ01015271 JAS40417.1 JAS47333.1 JAS54498.1 AXCN02000463 NNAY01000158 OXU30451.1 SPP77578.1 KRT08680.1 GEDC01016932 GEDC01014994 JAS20366.1 JAS22304.1 KRT08681.1 AAZX01002696 KZ288193 PBC34071.1 GBHO01013975 GBRD01013839 GDHC01019818 JAG29629.1 JAG51987.1 JAP98810.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000002320
UP000008820
+ More
UP000069940 UP000249989 UP000008792 UP000001070 UP000092553 UP000007798 UP000001292 UP000008711 UP000000803 UP000000304 UP000192221 UP000002282 UP000069272 UP000000673 UP000007801 UP000036403 UP000009192 UP000001819 UP000030765 UP000075920 UP000095301 UP000268350 UP000000311 UP000075880 UP000235965 UP000076408 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000075903 UP000007266 UP000075881 UP000075882 UP000075902 UP000075840 UP000076407 UP000075883 UP000075900 UP000005203 UP000008237 UP000078541 UP000053097 UP000279307 UP000078540 UP000005205 UP000095300 UP000007062 UP000078542 UP000075809 UP000019118 UP000030742 UP000078492 UP000007755 UP000091820 UP000008744 UP000075886 UP000075884 UP000215335 UP000002358 UP000075901 UP000075885 UP000242457
UP000069940 UP000249989 UP000008792 UP000001070 UP000092553 UP000007798 UP000001292 UP000008711 UP000000803 UP000000304 UP000192221 UP000002282 UP000069272 UP000000673 UP000007801 UP000036403 UP000009192 UP000001819 UP000030765 UP000075920 UP000095301 UP000268350 UP000000311 UP000075880 UP000235965 UP000076408 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000075903 UP000007266 UP000075881 UP000075882 UP000075902 UP000075840 UP000076407 UP000075883 UP000075900 UP000005203 UP000008237 UP000078541 UP000053097 UP000279307 UP000078540 UP000005205 UP000095300 UP000007062 UP000078542 UP000075809 UP000019118 UP000030742 UP000078492 UP000007755 UP000091820 UP000008744 UP000075886 UP000075884 UP000215335 UP000002358 UP000075901 UP000075885 UP000242457
Pfam
PF00850 Hist_deacetyl
Interpro
SUPFAM
SSF52768
SSF52768
Gene 3D
ProteinModelPortal
A0A2A4K903
A0A194PF89
A0A212F5S3
A0A0N1IGB9
A0A1Q3EXC7
B0WJ91
+ More
Q17CF0 A0A1Q3EXB1 A0A182GNN5 A0A023EUU4 B4MFY7 Q17CE9 A0A1Q3EXC6 U5EW58 B4IZ79 E3W281 E3W278 E3VX32 E3VX22 A0A0M4E8U0 B4MMY6 W8C8N5 A0A0J9RPK2 B4HU89 B3NC63 Q94517 B4QR08 A0A2M4AQV8 A0A1W4VAJ1 B4PIA1 A0A347ZJC4 A0A182FGH3 W5JF61 B3M9I7 A0A034WDK2 A0A0J7L4B5 A0A0A1WN83 B4KX99 A0A0K8VJ62 Q29EB4 A0A084WMI3 A0A182WBY2 A0A1Y1NG16 E3W282 S4NV65 A0A1I8M0Y6 A0A3B0JW31 A0A3B0J6G3 E2AL67 A0A182IKH6 A0A2J7QBD0 A0A336MS89 A0A182Y7V6 A0A1B0FJF6 A0A1A9Y199 A0A1A9UXP8 A0A1B0AZ17 A0A1A9ZWK4 A0A182V3V5 D6WG88 A0A182K1D6 A0A1L7N254 A0A182LKN9 A0A182TYI1 A0A182I495 A0A182X957 A0A182MWC0 A0A182RR67 A0A088AP92 E2B313 A0A195FTX4 A0A026WND6 A0A151I2N9 A0A158NKZ2 I1V501 A0A1I8PP67 Q7Q5D2 A0A151IM03 A0A151X3P9 J3JWV8 A0A151JAD5 F4WWT9 N6T0I9 A0A1L8DX14 A0A1A9W4R5 A0A1B6LWH4 B4HCY7 A0A1B6FBN9 A0A182QB58 A0A182NIY3 A0A232FI77 A0A3B0K2G2 A0A0R3P6D5 A0A1B6D3Z1 A0A0R3P695 K7J9J6 A0A182T387 A0A182NZT1 A0A2A3ESP9 A0A0A9YBV7
Q17CF0 A0A1Q3EXB1 A0A182GNN5 A0A023EUU4 B4MFY7 Q17CE9 A0A1Q3EXC6 U5EW58 B4IZ79 E3W281 E3W278 E3VX32 E3VX22 A0A0M4E8U0 B4MMY6 W8C8N5 A0A0J9RPK2 B4HU89 B3NC63 Q94517 B4QR08 A0A2M4AQV8 A0A1W4VAJ1 B4PIA1 A0A347ZJC4 A0A182FGH3 W5JF61 B3M9I7 A0A034WDK2 A0A0J7L4B5 A0A0A1WN83 B4KX99 A0A0K8VJ62 Q29EB4 A0A084WMI3 A0A182WBY2 A0A1Y1NG16 E3W282 S4NV65 A0A1I8M0Y6 A0A3B0JW31 A0A3B0J6G3 E2AL67 A0A182IKH6 A0A2J7QBD0 A0A336MS89 A0A182Y7V6 A0A1B0FJF6 A0A1A9Y199 A0A1A9UXP8 A0A1B0AZ17 A0A1A9ZWK4 A0A182V3V5 D6WG88 A0A182K1D6 A0A1L7N254 A0A182LKN9 A0A182TYI1 A0A182I495 A0A182X957 A0A182MWC0 A0A182RR67 A0A088AP92 E2B313 A0A195FTX4 A0A026WND6 A0A151I2N9 A0A158NKZ2 I1V501 A0A1I8PP67 Q7Q5D2 A0A151IM03 A0A151X3P9 J3JWV8 A0A151JAD5 F4WWT9 N6T0I9 A0A1L8DX14 A0A1A9W4R5 A0A1B6LWH4 B4HCY7 A0A1B6FBN9 A0A182QB58 A0A182NIY3 A0A232FI77 A0A3B0K2G2 A0A0R3P6D5 A0A1B6D3Z1 A0A0R3P695 K7J9J6 A0A182T387 A0A182NZT1 A0A2A3ESP9 A0A0A9YBV7
PDB
4BKX
E-value=0,
Score=1764
Ontologies
PATHWAY
GO
GO:0032041
GO:0005634
GO:0046872
GO:0004407
GO:1902692
GO:0000122
GO:0070983
GO:0070822
GO:0030261
GO:0035065
GO:0050771
GO:0008340
GO:0031523
GO:0007350
GO:0003714
GO:0005705
GO:0006342
GO:2001229
GO:0016581
GO:0035098
GO:0061086
GO:0045879
GO:0005700
GO:0048477
GO:0016580
GO:0010629
GO:0016575
GO:0008134
GO:0000118
GO:0006325
GO:0017053
GO:0006355
GO:0000785
GO:0016458
GO:0045664
GO:0045892
GO:0031061
GO:0005654
GO:0005737
GO:0070932
GO:0045944
GO:0070933
GO:0005840
GO:0006412
GO:0016705
GO:0020037
GO:0006811
GO:0016020
GO:0006260
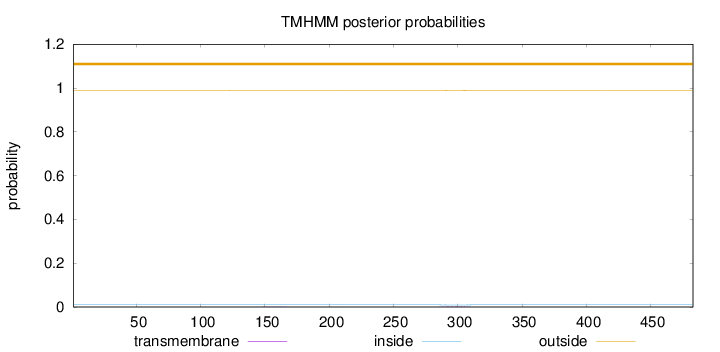
Topology
Subcellular location
Nucleus
Length:
483
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03981
Exp number, first 60 AAs:
0.0019
Total prob of N-in:
0.01053
outside
1 - 483
Population Genetic Test Statistics
Pi
192.435527
Theta
167.205133
Tajima's D
0.103474
CLR
1.240932
CSRT
0.39638018099095
Interpretation
Uncertain
