Gene
KWMTBOMO06729
Pre Gene Modal
BGIBMGA011744
Annotation
PREDICTED:_leucine-rich_repeat-containing_protein_16A_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.218 Peroxisomal Reliability : 1.053
Sequence
CDS
ATGCCAGCAAAATCACATATATCAACAGAGCTGAATGATAGCATCAGCAATGTTTTGGGGAAGAATGTCAAAGTTATTTATAAATCCCTTATTCGTATGGAATCAAGAGGTGATAAGGTTGATAACAAGGTTTTGGTTGTGACTGCATATCGGGTTTTTATAACTACAGCGAAAGTACCTACTAGGATTGACAATGGGTTCCATTTAATGGAAATAGAAGCCTTGGAGAGTAAAAAAGCGAATCATCTGTCCATAACATTGGTTCACGAACACAAGCCTGTCTCCATTCTCATAGGTGACGAAGGAACTTCAGAAGGTGTTATAAATGCCGTGAACGCTTTAACTGCCGCCTTCGATGATCTATTTCCTAATGTGCCAATGGTGGATATAATACAAAAGGTGAATCTACTCTTCCCATTACAAAACGTCAGCGGGACTAGGAAGCACCTAGCTTGTGGTGATTTTTCTAATCAGTACGCTGCCATGTGTGATTTATGTCAAACACCATTCAGAGCTGAAGTTGCGTGGGATATTGACACCATATACCTCGCACACGACATAAGAATGTTGCATTTGAAAGATTTCGATCATCTGGACCAAAGGGACCTGATACCACTAGTGATGGCGTTGCAACACAACACGTGGTTCGAGGGTATTTGCGGGGACGGGGTGCGGGCGAGTGGCGAGGCGTGGGAGAGCGTGTGCCGCGCGCTGCGGTGTTCGCGGCCGGCCACGCGCCGTGTCAGCTGGCGCAGCGCACAGCTCCGTCACGACCACGCCGCCAGACTGGGACACGCGCTCGCCCGCGCCCGCCACGCGCCCGCACTGCACACCATCGACTTCTCACAGAACCACATCGAGGATAAAGGTGCTATTAGTATACTGAGCGGCATATCGAACAATCCAGAAGGACTGAGGCACGTTGCGTTGTCACAATGCGGCGTGACGGGGAAGACGGTCGTACATTTGGCGACCATACTCAACGACAATCCCTGTCATCTCTCGACGCTCGCACACCTTGACCTGTCGCATAATAACCTGAAAGATGACGTGCACAATTTGTACAACTTCCTGGCACAGCCTAATGTATTGACCCATTTGAATTTGACTAACACGGAAACGACTTTAGAAAATATGTGGGGTGCCCTGCTGCGCGGGTGTGCGGCGCGCCTCACCTGGCTGTCCGTGTCCCGCAACCCGTGGTCGAGCGCCCGGCGCGCGCGGGACCCGCCGCCTTCCTTCCGACAATTCTTCACGGCGTGCCTCGCGATCAACCATCTCGACTTCTCCCACTGCAAAATGCCACCAGACGTGCTCAAGAGCTTGCTGCTGGGCCTGGCGTGCAACGAGAGTGCGGCCGGGGTCCAGCTGAATTTGTCGGGCGTGCTGAACTCGGCGCAGGCCGCGCACGTGCTCGAGTCGTGTATACACGGCGTGCGATGCCTGCAGTCTCTGGACCTCTCCGACAACAGTATGGAATTCGAATTGTCCGGTATTGTGAGAGCTATAGGGAAGAACCATTCCATCACACATTTATCGTTAAGTAGATTGACCGGGAAGAGATCGTACGCTCCCCCCCTAATACAAGCTCTAGTCCACGTGCTGCAGGAACCAGACACAGCGCTGACGTCACTGGACCTCAGCGATTGCAAACTAAAGGGTGATTTGTACGGATTATTGAACGCGTTGGGCGGAGCCCGGAGACTTCGTACACTGGACGTGGGAGGGAACCTAGCCGGCGACGCGGGAGCTAGATTACTTGCCAAAGCCTTACAGTGTAACTGTACTTTGCACACGCTGTATATAGATCGCAATGCATTTAGTTTACAAGGTTTGACTGACATTGCGACGGCGCTCCGTCGGTCTGTGAGCGTGCGGCGGGTCGAGTTCCCGGGATGCGACGCGGCGGCGGGCGGCCGCGCGGCCAGCGAGCGGGTCGCCGCGCTCTGGAGGGACTTGCACGAACGGTACTCGCCACTATTGTCACTCCGAACCACACGGATAGGTCCCTAA
Protein
MPAKSHISTELNDSISNVLGKNVKVIYKSLIRMESRGDKVDNKVLVVTAYRVFITTAKVPTRIDNGFHLMEIEALESKKANHLSITLVHEHKPVSILIGDEGTSEGVINAVNALTAAFDDLFPNVPMVDIIQKVNLLFPLQNVSGTRKHLACGDFSNQYAAMCDLCQTPFRAEVAWDIDTIYLAHDIRMLHLKDFDHLDQRDLIPLVMALQHNTWFEGICGDGVRASGEAWESVCRALRCSRPATRRVSWRSAQLRHDHAARLGHALARARHAPALHTIDFSQNHIEDKGAISILSGISNNPEGLRHVALSQCGVTGKTVVHLATILNDNPCHLSTLAHLDLSHNNLKDDVHNLYNFLAQPNVLTHLNLTNTETTLENMWGALLRGCAARLTWLSVSRNPWSSARRARDPPPSFRQFFTACLAINHLDFSHCKMPPDVLKSLLLGLACNESAAGVQLNLSGVLNSAQAAHVLESCIHGVRCLQSLDLSDNSMEFELSGIVRAIGKNHSITHLSLSRLTGKRSYAPPLIQALVHVLQEPDTALTSLDLSDCKLKGDLYGLLNALGGARRLRTLDVGGNLAGDAGARLLAKALQCNCTLHTLYIDRNAFSLQGLTDIATALRRSVSVRRVEFPGCDAAAGGRAASERVAALWRDLHERYSPLLSLRTTRIGP
Summary
Uniprot
A0A2A4KA86
A0A3S2M7S6
A0A0N0PCZ5
A0A194PEJ0
A0A212F5S6
H9JQD8
+ More
A0A1Y1LRL6 C6TP27 A0A067QY91 K7J5V8 A0A1I8N8B1 T1PLL0 A0A2P8XI95 A0A1Y1LRQ5 A0A1Y1LZB7 A0A1Y1LRS8 A0A1Y1LWA1 A0A0A1WZQ8 A0A0Q5WL40 A0A1W4VZ74 E1JGZ7 A0A0J9R7M1 A0A0R1DTJ3 A0A0P8YC22 A0A182RQG5 A0A0P9AF56 E2AID4 A0A0Q5WL52 A0A0A1XRS0 A0A1B6KVN4 A0A0Q5WNP1 B4J803 A0A1Q3FU64 A0A0K8VGJ4 B4LJH5 A0A1Q3FUN1 E1JGZ8 A0A0J9R8P1 A0A336K1P9 A0A336LQT9 A0A336LZY6 A0A0R1DN15 A0A0J9TVJ5 A0A336K803 A0A1I8N8B0 A0A3B0J2M8 A0A0K8VAL1 A0A0K8WA43 A0A1W4WBE9 B4GBF6 T1PLY5 A0A195F1W2 A0A1I8N8C9 A0A151JSX1 A0A0K8V7K3 A0A151XBK1 A0A034VBN4 A0A3B0JWJ1 W8ACX0 W8AJ71 A0A310SEB8 A0A087ZZK0 A0A195BH80 A0A0Q9XJN0 A0A158NL47 E2BKH2 A0A3B0JQ23 A0A195BYV6 A0A3B0J6J9 A0A3L8DAL1 Q7Q947 A0A1W4WA10 A0A0Q9W4M0 A0A026WW34 A0A3B0J2M7 A0A2A3EJE6 A0A1W4WAZ1 A0A0R3NMB2 A0A0P9BYS0 A0A0P8Y4L5 B3MHT1 A0A0J9R7M6 A0A0L7QNT5 A0A0R3NT91 W4VRD5 A0A084VUE3 A0A0Q5WC05 A0A1B6DKA9 B3N9V1 A0A0P4W380 A0A0Q9WCR9 A0A0A1WTD3 A0A0Q5WC88 A0A182WS46 A0A182HXU2 A0A1I8N8C1 A0A0K8V6G3 A0A0R1DM02
A0A1Y1LRL6 C6TP27 A0A067QY91 K7J5V8 A0A1I8N8B1 T1PLL0 A0A2P8XI95 A0A1Y1LRQ5 A0A1Y1LZB7 A0A1Y1LRS8 A0A1Y1LWA1 A0A0A1WZQ8 A0A0Q5WL40 A0A1W4VZ74 E1JGZ7 A0A0J9R7M1 A0A0R1DTJ3 A0A0P8YC22 A0A182RQG5 A0A0P9AF56 E2AID4 A0A0Q5WL52 A0A0A1XRS0 A0A1B6KVN4 A0A0Q5WNP1 B4J803 A0A1Q3FU64 A0A0K8VGJ4 B4LJH5 A0A1Q3FUN1 E1JGZ8 A0A0J9R8P1 A0A336K1P9 A0A336LQT9 A0A336LZY6 A0A0R1DN15 A0A0J9TVJ5 A0A336K803 A0A1I8N8B0 A0A3B0J2M8 A0A0K8VAL1 A0A0K8WA43 A0A1W4WBE9 B4GBF6 T1PLY5 A0A195F1W2 A0A1I8N8C9 A0A151JSX1 A0A0K8V7K3 A0A151XBK1 A0A034VBN4 A0A3B0JWJ1 W8ACX0 W8AJ71 A0A310SEB8 A0A087ZZK0 A0A195BH80 A0A0Q9XJN0 A0A158NL47 E2BKH2 A0A3B0JQ23 A0A195BYV6 A0A3B0J6J9 A0A3L8DAL1 Q7Q947 A0A1W4WA10 A0A0Q9W4M0 A0A026WW34 A0A3B0J2M7 A0A2A3EJE6 A0A1W4WAZ1 A0A0R3NMB2 A0A0P9BYS0 A0A0P8Y4L5 B3MHT1 A0A0J9R7M6 A0A0L7QNT5 A0A0R3NT91 W4VRD5 A0A084VUE3 A0A0Q5WC05 A0A1B6DKA9 B3N9V1 A0A0P4W380 A0A0Q9WCR9 A0A0A1WTD3 A0A0Q5WC88 A0A182WS46 A0A182HXU2 A0A1I8N8C1 A0A0K8V6G3 A0A0R1DM02
Pubmed
EMBL
NWSH01000021
PCG80703.1
RSAL01000016
RVE52984.1
KQ460499
KPJ14290.1
+ More
KQ459606 KPI91687.1 AGBW02010142 OWR49086.1 BABH01011901 BABH01011902 GEZM01048852 GEZM01048850 GEZM01048847 GEZM01048846 JAV76334.1 BT099513 ACU24753.2 KK852833 KDR15295.1 KA649667 AFP64296.1 PYGN01002038 PSN31727.1 GEZM01048848 JAV76339.1 GEZM01048853 JAV76327.1 GEZM01048843 JAV76353.1 GEZM01048842 JAV76355.1 GBXI01010000 JAD04292.1 CH954177 KQS70926.1 AE013599 ACZ94356.2 CM002911 KMY92066.1 CM000157 KRJ98307.1 CH902619 KPU76522.1 KPU76519.1 GL439757 EFN66809.1 KQS70923.1 GBXI01000620 JAD13672.1 GEBQ01024477 JAT15500.1 KQS70930.1 CH916367 EDW02233.1 GFDL01003884 JAV31161.1 GDHF01014341 JAI37973.1 CH940648 EDW61543.2 GFDL01003883 JAV31162.1 ACZ94354.1 KMY92069.1 UFQS01000063 UFQT01000063 SSW98884.1 SSX19270.1 SSW98885.1 SSX19271.1 SSW98882.1 SSX19268.1 KRJ98302.1 KMY92065.1 SSW98883.1 SSX19269.1 OUUW01000001 SPP75445.1 GDHF01016426 JAI35888.1 GDHF01004256 JAI48058.1 CH479181 EDW32258.1 KA649739 AFP64368.1 KQ981866 KYN34084.1 KQ978530 KYN30320.1 GDHF01017392 JAI34922.1 KQ982320 KYQ57734.1 GAKP01019023 JAC39929.1 SPP75448.1 GAMC01020775 JAB85780.1 GAMC01020773 JAB85782.1 KQ779356 OAD51968.1 KQ976488 KYM83526.1 CH933808 KRG04503.1 ADTU01019250 GL448795 EFN83812.1 SPP75446.1 KQ978501 KYM93505.1 SPP75442.1 QOIP01000011 RLU16958.1 AAAB01008905 EAA09715.3 KRF80057.1 KK107078 EZA60295.1 SPP75447.1 KZ288230 PBC31594.1 CM000071 KRT02071.1 KPU76523.1 KPU76526.1 EDV36918.2 KMY92063.1 KQ414855 KOC60151.1 KRT02064.1 GANO01003705 JAB56166.1 ATLV01016728 KE525103 KFB41587.1 KQS70928.1 GEDC01011230 JAS26068.1 EDV59647.2 GDRN01092799 JAI60041.1 KRF80055.1 GBXI01012003 JAD02289.1 KQS70924.1 APCN01005349 GDHF01017996 JAI34318.1 KRJ98308.1
KQ459606 KPI91687.1 AGBW02010142 OWR49086.1 BABH01011901 BABH01011902 GEZM01048852 GEZM01048850 GEZM01048847 GEZM01048846 JAV76334.1 BT099513 ACU24753.2 KK852833 KDR15295.1 KA649667 AFP64296.1 PYGN01002038 PSN31727.1 GEZM01048848 JAV76339.1 GEZM01048853 JAV76327.1 GEZM01048843 JAV76353.1 GEZM01048842 JAV76355.1 GBXI01010000 JAD04292.1 CH954177 KQS70926.1 AE013599 ACZ94356.2 CM002911 KMY92066.1 CM000157 KRJ98307.1 CH902619 KPU76522.1 KPU76519.1 GL439757 EFN66809.1 KQS70923.1 GBXI01000620 JAD13672.1 GEBQ01024477 JAT15500.1 KQS70930.1 CH916367 EDW02233.1 GFDL01003884 JAV31161.1 GDHF01014341 JAI37973.1 CH940648 EDW61543.2 GFDL01003883 JAV31162.1 ACZ94354.1 KMY92069.1 UFQS01000063 UFQT01000063 SSW98884.1 SSX19270.1 SSW98885.1 SSX19271.1 SSW98882.1 SSX19268.1 KRJ98302.1 KMY92065.1 SSW98883.1 SSX19269.1 OUUW01000001 SPP75445.1 GDHF01016426 JAI35888.1 GDHF01004256 JAI48058.1 CH479181 EDW32258.1 KA649739 AFP64368.1 KQ981866 KYN34084.1 KQ978530 KYN30320.1 GDHF01017392 JAI34922.1 KQ982320 KYQ57734.1 GAKP01019023 JAC39929.1 SPP75448.1 GAMC01020775 JAB85780.1 GAMC01020773 JAB85782.1 KQ779356 OAD51968.1 KQ976488 KYM83526.1 CH933808 KRG04503.1 ADTU01019250 GL448795 EFN83812.1 SPP75446.1 KQ978501 KYM93505.1 SPP75442.1 QOIP01000011 RLU16958.1 AAAB01008905 EAA09715.3 KRF80057.1 KK107078 EZA60295.1 SPP75447.1 KZ288230 PBC31594.1 CM000071 KRT02071.1 KPU76523.1 KPU76526.1 EDV36918.2 KMY92063.1 KQ414855 KOC60151.1 KRT02064.1 GANO01003705 JAB56166.1 ATLV01016728 KE525103 KFB41587.1 KQS70928.1 GEDC01011230 JAS26068.1 EDV59647.2 GDRN01092799 JAI60041.1 KRF80055.1 GBXI01012003 JAD02289.1 KQS70924.1 APCN01005349 GDHF01017996 JAI34318.1 KRJ98308.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
UP000005204
+ More
UP000027135 UP000002358 UP000095301 UP000245037 UP000008711 UP000192221 UP000000803 UP000002282 UP000007801 UP000075900 UP000000311 UP000001070 UP000008792 UP000268350 UP000008744 UP000078541 UP000078492 UP000075809 UP000005203 UP000078540 UP000009192 UP000005205 UP000008237 UP000078542 UP000279307 UP000007062 UP000053097 UP000242457 UP000001819 UP000053825 UP000030765 UP000076407 UP000075840
UP000027135 UP000002358 UP000095301 UP000245037 UP000008711 UP000192221 UP000000803 UP000002282 UP000007801 UP000075900 UP000000311 UP000001070 UP000008792 UP000268350 UP000008744 UP000078541 UP000078492 UP000075809 UP000005203 UP000078540 UP000009192 UP000005205 UP000008237 UP000078542 UP000279307 UP000007062 UP000053097 UP000242457 UP000001819 UP000053825 UP000030765 UP000076407 UP000075840
Interpro
Gene 3D
ProteinModelPortal
A0A2A4KA86
A0A3S2M7S6
A0A0N0PCZ5
A0A194PEJ0
A0A212F5S6
H9JQD8
+ More
A0A1Y1LRL6 C6TP27 A0A067QY91 K7J5V8 A0A1I8N8B1 T1PLL0 A0A2P8XI95 A0A1Y1LRQ5 A0A1Y1LZB7 A0A1Y1LRS8 A0A1Y1LWA1 A0A0A1WZQ8 A0A0Q5WL40 A0A1W4VZ74 E1JGZ7 A0A0J9R7M1 A0A0R1DTJ3 A0A0P8YC22 A0A182RQG5 A0A0P9AF56 E2AID4 A0A0Q5WL52 A0A0A1XRS0 A0A1B6KVN4 A0A0Q5WNP1 B4J803 A0A1Q3FU64 A0A0K8VGJ4 B4LJH5 A0A1Q3FUN1 E1JGZ8 A0A0J9R8P1 A0A336K1P9 A0A336LQT9 A0A336LZY6 A0A0R1DN15 A0A0J9TVJ5 A0A336K803 A0A1I8N8B0 A0A3B0J2M8 A0A0K8VAL1 A0A0K8WA43 A0A1W4WBE9 B4GBF6 T1PLY5 A0A195F1W2 A0A1I8N8C9 A0A151JSX1 A0A0K8V7K3 A0A151XBK1 A0A034VBN4 A0A3B0JWJ1 W8ACX0 W8AJ71 A0A310SEB8 A0A087ZZK0 A0A195BH80 A0A0Q9XJN0 A0A158NL47 E2BKH2 A0A3B0JQ23 A0A195BYV6 A0A3B0J6J9 A0A3L8DAL1 Q7Q947 A0A1W4WA10 A0A0Q9W4M0 A0A026WW34 A0A3B0J2M7 A0A2A3EJE6 A0A1W4WAZ1 A0A0R3NMB2 A0A0P9BYS0 A0A0P8Y4L5 B3MHT1 A0A0J9R7M6 A0A0L7QNT5 A0A0R3NT91 W4VRD5 A0A084VUE3 A0A0Q5WC05 A0A1B6DKA9 B3N9V1 A0A0P4W380 A0A0Q9WCR9 A0A0A1WTD3 A0A0Q5WC88 A0A182WS46 A0A182HXU2 A0A1I8N8C1 A0A0K8V6G3 A0A0R1DM02
A0A1Y1LRL6 C6TP27 A0A067QY91 K7J5V8 A0A1I8N8B1 T1PLL0 A0A2P8XI95 A0A1Y1LRQ5 A0A1Y1LZB7 A0A1Y1LRS8 A0A1Y1LWA1 A0A0A1WZQ8 A0A0Q5WL40 A0A1W4VZ74 E1JGZ7 A0A0J9R7M1 A0A0R1DTJ3 A0A0P8YC22 A0A182RQG5 A0A0P9AF56 E2AID4 A0A0Q5WL52 A0A0A1XRS0 A0A1B6KVN4 A0A0Q5WNP1 B4J803 A0A1Q3FU64 A0A0K8VGJ4 B4LJH5 A0A1Q3FUN1 E1JGZ8 A0A0J9R8P1 A0A336K1P9 A0A336LQT9 A0A336LZY6 A0A0R1DN15 A0A0J9TVJ5 A0A336K803 A0A1I8N8B0 A0A3B0J2M8 A0A0K8VAL1 A0A0K8WA43 A0A1W4WBE9 B4GBF6 T1PLY5 A0A195F1W2 A0A1I8N8C9 A0A151JSX1 A0A0K8V7K3 A0A151XBK1 A0A034VBN4 A0A3B0JWJ1 W8ACX0 W8AJ71 A0A310SEB8 A0A087ZZK0 A0A195BH80 A0A0Q9XJN0 A0A158NL47 E2BKH2 A0A3B0JQ23 A0A195BYV6 A0A3B0J6J9 A0A3L8DAL1 Q7Q947 A0A1W4WA10 A0A0Q9W4M0 A0A026WW34 A0A3B0J2M7 A0A2A3EJE6 A0A1W4WAZ1 A0A0R3NMB2 A0A0P9BYS0 A0A0P8Y4L5 B3MHT1 A0A0J9R7M6 A0A0L7QNT5 A0A0R3NT91 W4VRD5 A0A084VUE3 A0A0Q5WC05 A0A1B6DKA9 B3N9V1 A0A0P4W380 A0A0Q9WCR9 A0A0A1WTD3 A0A0Q5WC88 A0A182WS46 A0A182HXU2 A0A1I8N8C1 A0A0K8V6G3 A0A0R1DM02
PDB
4K17
E-value=2.21793e-95,
Score=893
Ontologies
GO
Topology
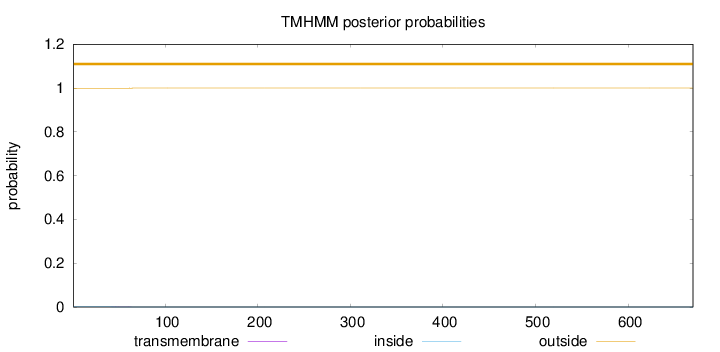
Length:
670
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01163
Exp number, first 60 AAs:
0.0098
Total prob of N-in:
0.00090
outside
1 - 670
Population Genetic Test Statistics
Pi
278.444956
Theta
182.252149
Tajima's D
1.576949
CLR
0.072757
CSRT
0.805459727013649
Interpretation
Uncertain
