Gene
KWMTBOMO06720
Annotation
PREDICTED:_intraflagellar_transport_protein_osm-1?_partial_[Papilio_polytes]
Full name
Intraflagellar transport protein 172 homolog
Alternative Name
Osmotic avoidance abnormal protein 1 homolog
Outer segment 2
Outer segment 2
Location in the cell
PlasmaMembrane Reliability : 3.798
Sequence
CDS
ATGAAGTTGGTGCTAATCGTGGGGTCGTGTGACGGTCGTTTGTCTGTGTATGAGGCGCAGCGCGGTGCCTTGTTGCGTAGTGTCGAGCCAGACTTGCCTCCGGACGTGAAAGATATCATAGCTGCGGCACCGAGTCCATCAGGACAGACTATTGCGTTTGGTGTCTTTAATGGATGTCTAGTGGGAGAGTTGAAGGAATCCGGGTCAGTCGATTTATCAATGCTTCAAATACCGAACCTGTACGCCACTAGAGCGCTCGCTTGGAGTAGTGACGGCACTAAATTGGCGATAGCTTCACAAACTGGAGCTCTTATAGTGCTCGAGGCTGTTTTAAGACGTTGGGTGTGGCGTGACACAGTGGAGGTGCAGCACGTGAGCTCGAGACAGTTGTTATTAACAAGACTAGCTAACGATGCTCTTCCGCTCACAGTTCTCACAAACCACGCTTCGGATATATTCGGCGTCCGTTTCATTGGGAATGACTGGTATGCGGTGTGCAGGACTAGCAACACGTTGATCTTATGCGATATAGCTCGTGGACTCACCAGTGAGATACTATGGACGGGTAGCGGAGAACGTATTTATGCAGCTGTCGGTGGAGCATGTCTGATATATCGCGCCGGTGAACTGAGTGTTGTCGAATACGGACTTGATAAAGTTCTACATACGGTTCGTACAGAACGTGTGAACCCTCACGTGATCAGCGTGCGGATCAATGAAGCTGGTTTGACGAAAGGGGACTGCAAATACTTGGCATATTTGCTAGATAGACAAACTATTGCTGTTGTAGATTTGAGCACAGGTGCTCAGTTGGGGCAGTGGTGGCACGAGGCCCGCGTGGATTGGCTCGAGCTGAACGAGACCGCGCAGCTGTTGCTGCTCCGTGACACGAGGAGAAGACTTGCGCTACTGCGGTTACCGTCTGGTGATAAGGAAATTATTGCGAGTGGCGTCTGGCCTAAAGATAATGACACTCTTATTGCTCTACAACGTACATATTTTACATTTATATCTATCATTATTTTTGTTATCATGAAACGTTAG
Protein
MKLVLIVGSCDGRLSVYEAQRGALLRSVEPDLPPDVKDIIAAAPSPSGQTIAFGVFNGCLVGELKESGSVDLSMLQIPNLYATRALAWSSDGTKLAIASQTGALIVLEAVLRRWVWRDTVEVQHVSSRQLLLTRLANDALPLTVLTNHASDIFGVRFIGNDWYAVCRTSNTLILCDIARGLTSEILWTGSGERIYAAVGGACLIYRAGELSVVEYGLDKVLHTVRTERVNPHVISVRINEAGLTKGDCKYLAYLLDRQTIAVVDLSTGAQLGQWWHEARVDWLELNETAQLLLLRDTRRRLALLRLPSGDKEIIASGVWPKDNDTLIALQRTYFTFISIIIFVIMKR
Summary
Description
Required for the maintenance and formation of cilia.
Similarity
Belongs to the IFT172 family.
Keywords
Cell projection
Cilium
Complete proteome
Developmental protein
Reference proteome
Repeat
TPR repeat
WD repeat
Feature
chain Intraflagellar transport protein 172 homolog
Uniprot
A0A2H1W743
A0A3S2NAW4
A0A2A4K9V4
A0A0N1PI26
A0A194PG50
A0A212EVD8
+ More
B4QN35 A0A0T6AZB0 B4N307 B4PEC2 A0A182T8W3 B3NBF4 A0A1B0CTA0 A0A0L0CF00 B4HW57 A0A1W4W4F3 A0A0R3P5G5 Q2LYU0 A0A1I8PPJ7 A0A0J9RMB7 A0A0J9RMU1 Q9W040 A0A1J1I5Q0 A0A182ILK1 A0A1I8NB59 A0A3B0KUL4 B0W088 A0A084VAU0 B3M755 A0A182YPZ2 A0A336LY03 A0A182PGF5 A0A1A9XHG5 A0A182UJV6 A0A182VL23 A0A182FHA6 A0A182HMH4 A0A1A9W7J7 A0A182RX30 A0A182WAJ7 Q5TRR4 A0A182JUZ2 A0A182MI61 A0A182N7Y3 A0A182QPT3 B4KV02 A0A1A9VEC2 A0A1A9Z4J4 A0A1B0F9U2 A0A1S4F9Q5 Q17B06 Q17AW7 A0A0C9QDS5 A0A1B0AKI5 A0A0M4EC07 A0A1B0BFI1 A0A182XD40 B4LBX2 E0VLV6 D7EJZ7 A0A0K2TUP4 B4J0J1 A0A1B6ET01 A0A2J7R9T7 A0A224XHN2 W5JIP2 T1HGF8 A0A0J7K4F0 A0A1B6CK41 A0A1B6D0F4 A0A1B6EBJ3 A0A067RFG0 E2AAT0 A0A1A8L5D0 A0A232FA46 K7IXX9 N6TQB8 U4UJF4 A0A3L8DJV9 A0A1A8QEB1 A0A026WHM9 Q5FWQ3 A0A2K6G5Y4 H3CIP1 W5KZE2 A0A1L8GCV5 F6SG36 A0A3B4BQL3 A0A3B4BQU6 A0A3B4BLU9 B3DK94 A0A3B4BQG8 A0A2S2QPL8 A0A2K5WX28 V9K8U8 Q5RHH4 A0A0P7V0V5 A0A1W4YW35 A0A1W4YLQ0 A0A3B3T3K1
B4QN35 A0A0T6AZB0 B4N307 B4PEC2 A0A182T8W3 B3NBF4 A0A1B0CTA0 A0A0L0CF00 B4HW57 A0A1W4W4F3 A0A0R3P5G5 Q2LYU0 A0A1I8PPJ7 A0A0J9RMB7 A0A0J9RMU1 Q9W040 A0A1J1I5Q0 A0A182ILK1 A0A1I8NB59 A0A3B0KUL4 B0W088 A0A084VAU0 B3M755 A0A182YPZ2 A0A336LY03 A0A182PGF5 A0A1A9XHG5 A0A182UJV6 A0A182VL23 A0A182FHA6 A0A182HMH4 A0A1A9W7J7 A0A182RX30 A0A182WAJ7 Q5TRR4 A0A182JUZ2 A0A182MI61 A0A182N7Y3 A0A182QPT3 B4KV02 A0A1A9VEC2 A0A1A9Z4J4 A0A1B0F9U2 A0A1S4F9Q5 Q17B06 Q17AW7 A0A0C9QDS5 A0A1B0AKI5 A0A0M4EC07 A0A1B0BFI1 A0A182XD40 B4LBX2 E0VLV6 D7EJZ7 A0A0K2TUP4 B4J0J1 A0A1B6ET01 A0A2J7R9T7 A0A224XHN2 W5JIP2 T1HGF8 A0A0J7K4F0 A0A1B6CK41 A0A1B6D0F4 A0A1B6EBJ3 A0A067RFG0 E2AAT0 A0A1A8L5D0 A0A232FA46 K7IXX9 N6TQB8 U4UJF4 A0A3L8DJV9 A0A1A8QEB1 A0A026WHM9 Q5FWQ3 A0A2K6G5Y4 H3CIP1 W5KZE2 A0A1L8GCV5 F6SG36 A0A3B4BQL3 A0A3B4BQU6 A0A3B4BLU9 B3DK94 A0A3B4BQG8 A0A2S2QPL8 A0A2K5WX28 V9K8U8 Q5RHH4 A0A0P7V0V5 A0A1W4YW35 A0A1W4YLQ0 A0A3B3T3K1
Pubmed
26354079
22118469
17994087
17550304
26108605
15632085
+ More
22936249 10731132 12537572 25315136 24438588 25244985 12364791 14747013 17210077 17510324 20566863 18362917 19820115 20920257 23761445 24845553 20798317 28648823 20075255 23537049 30249741 24508170 15496914 25329095 27762356 20431018 24402279 15269167 23594743 24140113 29240929
22936249 10731132 12537572 25315136 24438588 25244985 12364791 14747013 17210077 17510324 20566863 18362917 19820115 20920257 23761445 24845553 20798317 28648823 20075255 23537049 30249741 24508170 15496914 25329095 27762356 20431018 24402279 15269167 23594743 24140113 29240929
EMBL
ODYU01006762
SOQ48901.1
RSAL01000137
RVE46188.1
NWSH01000021
PCG80678.1
+ More
KQ460499 KPJ14298.1 KQ459606 KPI91679.1 AGBW02012204 OWR45456.1 CM000363 EDX08917.1 LJIG01022553 KRT79959.1 CH964062 EDW78746.1 CM000159 EDW92960.1 CH954178 EDV50276.1 AJWK01027318 AJWK01027319 AJWK01027320 AJWK01027321 JRES01000493 KNC30792.1 CH480817 EDW50172.1 CH379069 KRT08360.1 EAL29770.2 CM002912 KMY97056.1 KMY97058.1 KMY97057.1 AE014296 CVRI01000042 CRK95655.1 OUUW01000012 SPP87638.1 DS231817 EDS39627.1 ATLV01004460 KE524214 KFB35084.1 CH902618 EDV38716.1 UFQS01000198 UFQT01000198 SSX01209.1 SSX21589.1 APCN01004435 AAAB01008960 EAL39797.1 AXCM01002609 AXCN02000907 CH933809 EDW18313.1 CCAG010018360 CH477327 EAT43464.1 CH477329 EAT43404.1 GBYB01001479 JAG71246.1 JXJN01025783 CP012525 ALC42884.1 JXJN01013485 CH940647 EDW69772.1 DS235281 EEB14362.1 DS497723 EFA12908.1 HACA01012179 CDW29540.1 CH916366 EDV96827.1 GECZ01028721 JAS41048.1 NEVH01006580 PNF37590.1 GFTR01008877 JAW07549.1 ADMH02001057 ETN64252.1 ACPB03006864 LBMM01014452 KMQ85174.1 GEDC01023434 JAS13864.1 GEDC01018223 JAS19075.1 GEDC01002054 JAS35244.1 KK852721 KDR17746.1 GL438172 EFN69459.1 HAEF01002628 SBR40010.1 NNAY01000552 OXU27721.1 APGK01055682 APGK01055683 KB741269 ENN71435.1 KB632330 ERL92593.1 QOIP01000007 RLU20158.1 HAEH01011209 SBR91663.1 KK107238 EZA54589.1 BC089250 AAH89250.1 CM004474 OCT81621.1 AAMC01141121 AAMC01141122 AAMC01141123 AAMC01141124 AAMC01141125 BC163767 AAI63767.1 GGMS01010267 MBY79470.1 AQIA01018728 AQIA01018729 AQIA01018730 AQIA01018731 JW861520 AFO94037.1 AY618923 BX511178 JARO02001718 KPP74606.1
KQ460499 KPJ14298.1 KQ459606 KPI91679.1 AGBW02012204 OWR45456.1 CM000363 EDX08917.1 LJIG01022553 KRT79959.1 CH964062 EDW78746.1 CM000159 EDW92960.1 CH954178 EDV50276.1 AJWK01027318 AJWK01027319 AJWK01027320 AJWK01027321 JRES01000493 KNC30792.1 CH480817 EDW50172.1 CH379069 KRT08360.1 EAL29770.2 CM002912 KMY97056.1 KMY97058.1 KMY97057.1 AE014296 CVRI01000042 CRK95655.1 OUUW01000012 SPP87638.1 DS231817 EDS39627.1 ATLV01004460 KE524214 KFB35084.1 CH902618 EDV38716.1 UFQS01000198 UFQT01000198 SSX01209.1 SSX21589.1 APCN01004435 AAAB01008960 EAL39797.1 AXCM01002609 AXCN02000907 CH933809 EDW18313.1 CCAG010018360 CH477327 EAT43464.1 CH477329 EAT43404.1 GBYB01001479 JAG71246.1 JXJN01025783 CP012525 ALC42884.1 JXJN01013485 CH940647 EDW69772.1 DS235281 EEB14362.1 DS497723 EFA12908.1 HACA01012179 CDW29540.1 CH916366 EDV96827.1 GECZ01028721 JAS41048.1 NEVH01006580 PNF37590.1 GFTR01008877 JAW07549.1 ADMH02001057 ETN64252.1 ACPB03006864 LBMM01014452 KMQ85174.1 GEDC01023434 JAS13864.1 GEDC01018223 JAS19075.1 GEDC01002054 JAS35244.1 KK852721 KDR17746.1 GL438172 EFN69459.1 HAEF01002628 SBR40010.1 NNAY01000552 OXU27721.1 APGK01055682 APGK01055683 KB741269 ENN71435.1 KB632330 ERL92593.1 QOIP01000007 RLU20158.1 HAEH01011209 SBR91663.1 KK107238 EZA54589.1 BC089250 AAH89250.1 CM004474 OCT81621.1 AAMC01141121 AAMC01141122 AAMC01141123 AAMC01141124 AAMC01141125 BC163767 AAI63767.1 GGMS01010267 MBY79470.1 AQIA01018728 AQIA01018729 AQIA01018730 AQIA01018731 JW861520 AFO94037.1 AY618923 BX511178 JARO02001718 KPP74606.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
UP000000304
+ More
UP000007798 UP000002282 UP000075901 UP000008711 UP000092461 UP000037069 UP000001292 UP000192221 UP000001819 UP000095300 UP000000803 UP000183832 UP000075880 UP000095301 UP000268350 UP000002320 UP000030765 UP000007801 UP000076408 UP000075885 UP000092443 UP000075902 UP000075903 UP000069272 UP000075840 UP000091820 UP000075900 UP000075920 UP000007062 UP000075881 UP000075883 UP000075884 UP000075886 UP000009192 UP000078200 UP000092445 UP000092444 UP000008820 UP000092460 UP000092553 UP000076407 UP000008792 UP000009046 UP000007266 UP000001070 UP000235965 UP000000673 UP000015103 UP000036403 UP000027135 UP000000311 UP000215335 UP000002358 UP000019118 UP000030742 UP000279307 UP000053097 UP000233160 UP000007303 UP000018467 UP000186698 UP000008143 UP000261440 UP000233100 UP000000437 UP000034805 UP000192224 UP000261540
UP000007798 UP000002282 UP000075901 UP000008711 UP000092461 UP000037069 UP000001292 UP000192221 UP000001819 UP000095300 UP000000803 UP000183832 UP000075880 UP000095301 UP000268350 UP000002320 UP000030765 UP000007801 UP000076408 UP000075885 UP000092443 UP000075902 UP000075903 UP000069272 UP000075840 UP000091820 UP000075900 UP000075920 UP000007062 UP000075881 UP000075883 UP000075884 UP000075886 UP000009192 UP000078200 UP000092445 UP000092444 UP000008820 UP000092460 UP000092553 UP000076407 UP000008792 UP000009046 UP000007266 UP000001070 UP000235965 UP000000673 UP000015103 UP000036403 UP000027135 UP000000311 UP000215335 UP000002358 UP000019118 UP000030742 UP000279307 UP000053097 UP000233160 UP000007303 UP000018467 UP000186698 UP000008143 UP000261440 UP000233100 UP000000437 UP000034805 UP000192224 UP000261540
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR001680 WD40_repeat
IPR011047 Quinoprotein_ADH-like_supfam
IPR017986 WD40_repeat_dom
IPR007714 CFA20_dom
IPR036396 Cyt_P450_sf
IPR024977 Apc4_WD40_dom
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR002401 Cyt_P450_E_grp-I
IPR019734 TPR_repeat
IPR016024 ARM-type_fold
IPR011044 Quino_amine_DH_bsu
IPR011048 Haem_d1_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR001680 WD40_repeat
IPR011047 Quinoprotein_ADH-like_supfam
IPR017986 WD40_repeat_dom
IPR007714 CFA20_dom
IPR036396 Cyt_P450_sf
IPR024977 Apc4_WD40_dom
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR002401 Cyt_P450_E_grp-I
IPR019734 TPR_repeat
IPR016024 ARM-type_fold
IPR011044 Quino_amine_DH_bsu
IPR011048 Haem_d1_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1W743
A0A3S2NAW4
A0A2A4K9V4
A0A0N1PI26
A0A194PG50
A0A212EVD8
+ More
B4QN35 A0A0T6AZB0 B4N307 B4PEC2 A0A182T8W3 B3NBF4 A0A1B0CTA0 A0A0L0CF00 B4HW57 A0A1W4W4F3 A0A0R3P5G5 Q2LYU0 A0A1I8PPJ7 A0A0J9RMB7 A0A0J9RMU1 Q9W040 A0A1J1I5Q0 A0A182ILK1 A0A1I8NB59 A0A3B0KUL4 B0W088 A0A084VAU0 B3M755 A0A182YPZ2 A0A336LY03 A0A182PGF5 A0A1A9XHG5 A0A182UJV6 A0A182VL23 A0A182FHA6 A0A182HMH4 A0A1A9W7J7 A0A182RX30 A0A182WAJ7 Q5TRR4 A0A182JUZ2 A0A182MI61 A0A182N7Y3 A0A182QPT3 B4KV02 A0A1A9VEC2 A0A1A9Z4J4 A0A1B0F9U2 A0A1S4F9Q5 Q17B06 Q17AW7 A0A0C9QDS5 A0A1B0AKI5 A0A0M4EC07 A0A1B0BFI1 A0A182XD40 B4LBX2 E0VLV6 D7EJZ7 A0A0K2TUP4 B4J0J1 A0A1B6ET01 A0A2J7R9T7 A0A224XHN2 W5JIP2 T1HGF8 A0A0J7K4F0 A0A1B6CK41 A0A1B6D0F4 A0A1B6EBJ3 A0A067RFG0 E2AAT0 A0A1A8L5D0 A0A232FA46 K7IXX9 N6TQB8 U4UJF4 A0A3L8DJV9 A0A1A8QEB1 A0A026WHM9 Q5FWQ3 A0A2K6G5Y4 H3CIP1 W5KZE2 A0A1L8GCV5 F6SG36 A0A3B4BQL3 A0A3B4BQU6 A0A3B4BLU9 B3DK94 A0A3B4BQG8 A0A2S2QPL8 A0A2K5WX28 V9K8U8 Q5RHH4 A0A0P7V0V5 A0A1W4YW35 A0A1W4YLQ0 A0A3B3T3K1
B4QN35 A0A0T6AZB0 B4N307 B4PEC2 A0A182T8W3 B3NBF4 A0A1B0CTA0 A0A0L0CF00 B4HW57 A0A1W4W4F3 A0A0R3P5G5 Q2LYU0 A0A1I8PPJ7 A0A0J9RMB7 A0A0J9RMU1 Q9W040 A0A1J1I5Q0 A0A182ILK1 A0A1I8NB59 A0A3B0KUL4 B0W088 A0A084VAU0 B3M755 A0A182YPZ2 A0A336LY03 A0A182PGF5 A0A1A9XHG5 A0A182UJV6 A0A182VL23 A0A182FHA6 A0A182HMH4 A0A1A9W7J7 A0A182RX30 A0A182WAJ7 Q5TRR4 A0A182JUZ2 A0A182MI61 A0A182N7Y3 A0A182QPT3 B4KV02 A0A1A9VEC2 A0A1A9Z4J4 A0A1B0F9U2 A0A1S4F9Q5 Q17B06 Q17AW7 A0A0C9QDS5 A0A1B0AKI5 A0A0M4EC07 A0A1B0BFI1 A0A182XD40 B4LBX2 E0VLV6 D7EJZ7 A0A0K2TUP4 B4J0J1 A0A1B6ET01 A0A2J7R9T7 A0A224XHN2 W5JIP2 T1HGF8 A0A0J7K4F0 A0A1B6CK41 A0A1B6D0F4 A0A1B6EBJ3 A0A067RFG0 E2AAT0 A0A1A8L5D0 A0A232FA46 K7IXX9 N6TQB8 U4UJF4 A0A3L8DJV9 A0A1A8QEB1 A0A026WHM9 Q5FWQ3 A0A2K6G5Y4 H3CIP1 W5KZE2 A0A1L8GCV5 F6SG36 A0A3B4BQL3 A0A3B4BQU6 A0A3B4BLU9 B3DK94 A0A3B4BQG8 A0A2S2QPL8 A0A2K5WX28 V9K8U8 Q5RHH4 A0A0P7V0V5 A0A1W4YW35 A0A1W4YLQ0 A0A3B3T3K1
Ontologies
KEGG
GO
GO:0031514
GO:0042073
GO:0097730
GO:1905515
GO:0005929
GO:0007018
GO:0060271
GO:0030990
GO:0005871
GO:0007275
GO:0020037
GO:0005506
GO:0016705
GO:0005930
GO:0036064
GO:0030992
GO:0045494
GO:0032006
GO:0060041
GO:0048793
GO:0005515
GO:0008270
GO:0043565
GO:0008242
GO:0006811
GO:0016020
GO:0005634
GO:0006260
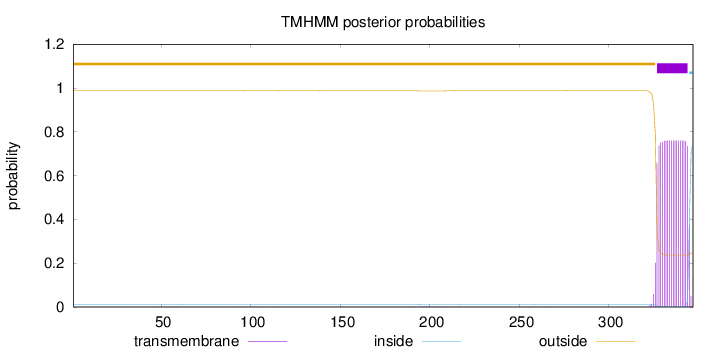
Topology
Subcellular location
Length:
347
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
14.2705
Exp number, first 60 AAs:
0.00488
Total prob of N-in:
0.01215
outside
1 - 326
TMhelix
327 - 344
inside
345 - 347
Population Genetic Test Statistics
Pi
250.089579
Theta
216.383768
Tajima's D
0.526412
CLR
0.191857
CSRT
0.51732413379331
Interpretation
Uncertain
