Pre Gene Modal
BGIBMGA011750
Annotation
PREDICTED:_intraflagellar_transport_protein_osm-1_[Bombyx_mori]
Full name
Intraflagellar transport protein 172 homolog
Alternative Name
Osmotic avoidance abnormal protein 1 homolog
Outer segment 2
Outer segment 2
Location in the cell
Cytoplasmic Reliability : 1.64
Sequence
CDS
ATGGGTTGTGCCAAGTATTTAGACGGCGTAACGAACAGCGCTGACGTCGCACCGCTCTGGAGGCAGCTGGCCGAACGAGCTCTGCAAAATGATGATATACAGTTAGCTGCAAAGTGTTATCGAGAAATAGGCGATGAATCCAGAACATTTTATTTGGAGAAGATTATAGAAGCTGCTGCGGCTGAAGGCGATGGAGACGTCGCTGCAGGTCTGAGCAGCGCAGAAGTTAGGGCACGATTAGCAATATTCAATGGGGACCTAGCGACGGCCGAAGCCTGGTATGCGAGACGATCCGGTCACCCTGAACGCGCCATCGCCATGTATAAGCAGTTCAACAGGTGGACTGACGCTATTGCTCTGGCCGAGAAGGCGGATCGCACTTCAGTTGCAAATTTAAAGCAACAGTACATGGATTATTTAACCTCAACTGGTCGTTTGGGAGAGGCCGGGGCTACCCTGGCAGCTGCAGGAGATGTACAGGGCGCAGTGAGACTGTGGTTGCGAGGAGGAAGGGCTCGTAGAGCTGCCTCACTCCTGTTACAGCATCCACAGCTACTGAGGGATAATGAGTTACTAGACTCTGTGCACACACAACTCATACAGGAGGAATGGTGGGAAATGGCCGGTGAATTATCTGAGAGACGAGGCGATGTCCGTTTGGCTGTAGATTATTACGCTAAAGGAAATAATTTTGCCAGAGCCGTTCAACTTGCCAGAGAGTCTTGTCCGGGTGAAGTGACGCATTTGGAAGGGCAGTGGGGCGCGTGGCTGGTGTCGTCCCGCCAGGCCGGCGCCGCCGTGCCGCACCTCATCGAGGCGGGGGACACTCGCGCTGCGCTCGCGGCGGCTATCAAGGCGCACCATTACAGAAAAGCTCTTCAGATTGTGCAGGCCATAGATGATAAGGAATCTATAAAAGAACAATGCGAGCAGCTCGGTGATCATTTAATATCTACGCAGGAATGGGAGACAGCAGAGAAGTTTCTCACTTCATGCGGTATGGCGGAGCGTTGCGTTCGAGCATACAACGGGGTCGGTCGCTTCGCGGACGGCCTCAGGCTTGCGGCTTCACAACTGACGGAGGAACAAACGCGAGACATTTATATACCGCTGGCCGAGGAGCTGAGGAAAGACGGACAACTTAGAAAAGCCGAGCAAATTTATATAGGTCTAGGAGAGGCTGATGAAGCTATTTCAATGTACAAGGAGGCGTCTCAATACGAGTCAATGTTGCGTCTGGTCGCGATACACCGATCCACTTTGTTAGAGGCGACGCGTCGACACGTTGCCCAAGCGTTACACGCCTCGGGAGATTTACGTGCGGCTGAAAACTATTATATACAAGCCGGTGACTGGAAAAGTGCCATACAGATGTATCGTTCGGCCGCACAGTGGGAGGGTGCTGAGCGTGTTGCTAGAGCACACGCTCCTGTTAATGTACAACAGCAGGTGGCGCTACAATGGGCCGCTGTTTTGGGTGGATTGGCTGGTGCACGACTTCTGGCTGCAAGAGGATTGGCTGCTTTAGGAGCACGATGGGCCCTACAGGCACAAAGATGGGATATAGCAATAGAATTGAGCGAACTCGGTGGTGCTATCACTCGGCGTGAAGTGGCTCGACACGAAGCGGCTGCGTTGGCGGAGGAACATCCTGAGGAGGCTGAAGCAGCCTTCCTTCGCGCGTCGGCTCCCGACCACGCCGTGCGCATGTGGCTCTCTAAAGAACAACACCAGAGGGCGCTGATCTTAGCCGAGCAGCATGCGCCGCATATGGTCGAAGAAGTACTAGTTGAAGGCGCTAGAGCAGCCGCAGAGAGAGGCGATTTACAGCAGTTCGAAGTACTCATGATCCGAGCTAATAGGCCGAGAGAGGTCGTCCAGCACTACAAGGATCTAGAGTTATGGGAAGAGGCTGCCCGTGTGTCTCGTGAGTATTTGCCGTCTAGCGGCGACGAGGAAGACGTGCCACCGGCCGTGCCAGCCCTGCTGACGCAAGCCGCGGCTCACGCCGACGCCGGCGAGTGGTGGGAAGCCGTGCAGGTATCGTATACTCAGAGTGTCTACAGGTTAAATTCAGTGGTGGCTGAGGCAATGCGGTGCCTGATTTTCGCAATTTCATAA
Protein
MGCAKYLDGVTNSADVAPLWRQLAERALQNDDIQLAAKCYREIGDESRTFYLEKIIEAAAAEGDGDVAAGLSSAEVRARLAIFNGDLATAEAWYARRSGHPERAIAMYKQFNRWTDAIALAEKADRTSVANLKQQYMDYLTSTGRLGEAGATLAAAGDVQGAVRLWLRGGRARRAASLLLQHPQLLRDNELLDSVHTQLIQEEWWEMAGELSERRGDVRLAVDYYAKGNNFARAVQLARESCPGEVTHLEGQWGAWLVSSRQAGAAVPHLIEAGDTRAALAAAIKAHHYRKALQIVQAIDDKESIKEQCEQLGDHLISTQEWETAEKFLTSCGMAERCVRAYNGVGRFADGLRLAASQLTEEQTRDIYIPLAEELRKDGQLRKAEQIYIGLGEADEAISMYKEASQYESMLRLVAIHRSTLLEATRRHVAQALHASGDLRAAENYYIQAGDWKSAIQMYRSAAQWEGAERVARAHAPVNVQQQVALQWAAVLGGLAGARLLAARGLAALGARWALQAQRWDIAIELSELGGAITRREVARHEAAALAEEHPEEAEAAFLRASAPDHAVRMWLSKEQHQRALILAEQHAPHMVEEVLVEGARAAAERGDLQQFEVLMIRANRPREVVQHYKDLELWEEAARVSREYLPSSGDEEDVPPAVPALLTQAAAHADAGEWWEAVQVSYTQSVYRLNSVVAEAMRCLIFAIS
Summary
Description
Required for the maintenance and formation of cilia.
Similarity
Belongs to the IFT172 family.
Keywords
Cell projection
Cilium
Complete proteome
Developmental protein
Reference proteome
Repeat
TPR repeat
WD repeat
Feature
chain Intraflagellar transport protein 172 homolog
Uniprot
H9JQE4
A0A3S2NAW4
A0A2A4K9V4
A0A2H1W743
A0A194PG50
A0A1S4F9Q5
+ More
Q17B06 A0A1W4W4F3 Q17AW7 Q9W040 B4HW57 A0A0J9RMB7 A0A0J9RMU1 B0W088 B3NBF4 B4LBX2 A0A182PGF5 A0A3B0KUL4 B4PEC2 A0A182QPT3 A0A182JUZ2 B3M755 B4N307 A0A1A9Z4J4 B4KV02 A0A1B0F9U2 A0A1B0AKI5 A0A1A9VEC2 A0A1B0BFI1 A0A182XD40 A0A182UJV6 Q5TRR4 A0A182RX30 A0A182HMH4 A0A182VL23 A0A084VAU0 Q2LYU0 B4J0J1 A0A182YPZ2 A0A0R3P5G5 A0A182WAJ7 A0A182MI61 A0A182N7Y3 T1ISG8 A0A1A9W7J7 A0A1I8NB59 A0A182FHA6 A0A0L0CF00 A0A1J1I5Q0 D7EJZ7 A0A1I8PPJ7 W5JIP2 A0A131Y0L6 A0A147BMA5 A0A067RFG0 A0A0V1GW14 A0A0V1N8M8 A0A0K2TVY2 A0A0V1N7V3 A0A0V1N841 E0VLV6 B7Q8U1 A0A0V1GVW5 A0A0V0TP49 A0A0V1GVJ3 A0A0V1GW08 A0A0V1GVV6 A0A0V1AG17 A0A0V1AG32 N6TQB8 A0A0V0X6A3 A0A0V1NVW4 A0A0V1LV56 J9L180 A0A224XHN2 A0A0M4EC07 A0A0N5E211 A0A0V1BWV4 A0A0V1D0D9 A0A0V1D0C5 W5U640 U4UJF4 A0A2S2QQN7 A0A3B4BQU6 A0A3B4BQG8 A0A3B4BLU9 A0A3Q1AR83 A0A3P8SZX4 A0A1B6EBJ3 A0A1B6D0F4 A0A210PPZ5 B3DK94 A0A1B6CK41 A0A1W4YLQ0 A0A1W4YW35 A0A1X7UFI5 A0A3B4BGA9 Q5RHH4
Q17B06 A0A1W4W4F3 Q17AW7 Q9W040 B4HW57 A0A0J9RMB7 A0A0J9RMU1 B0W088 B3NBF4 B4LBX2 A0A182PGF5 A0A3B0KUL4 B4PEC2 A0A182QPT3 A0A182JUZ2 B3M755 B4N307 A0A1A9Z4J4 B4KV02 A0A1B0F9U2 A0A1B0AKI5 A0A1A9VEC2 A0A1B0BFI1 A0A182XD40 A0A182UJV6 Q5TRR4 A0A182RX30 A0A182HMH4 A0A182VL23 A0A084VAU0 Q2LYU0 B4J0J1 A0A182YPZ2 A0A0R3P5G5 A0A182WAJ7 A0A182MI61 A0A182N7Y3 T1ISG8 A0A1A9W7J7 A0A1I8NB59 A0A182FHA6 A0A0L0CF00 A0A1J1I5Q0 D7EJZ7 A0A1I8PPJ7 W5JIP2 A0A131Y0L6 A0A147BMA5 A0A067RFG0 A0A0V1GW14 A0A0V1N8M8 A0A0K2TVY2 A0A0V1N7V3 A0A0V1N841 E0VLV6 B7Q8U1 A0A0V1GVW5 A0A0V0TP49 A0A0V1GVJ3 A0A0V1GW08 A0A0V1GVV6 A0A0V1AG17 A0A0V1AG32 N6TQB8 A0A0V0X6A3 A0A0V1NVW4 A0A0V1LV56 J9L180 A0A224XHN2 A0A0M4EC07 A0A0N5E211 A0A0V1BWV4 A0A0V1D0D9 A0A0V1D0C5 W5U640 U4UJF4 A0A2S2QQN7 A0A3B4BQU6 A0A3B4BQG8 A0A3B4BLU9 A0A3Q1AR83 A0A3P8SZX4 A0A1B6EBJ3 A0A1B6D0F4 A0A210PPZ5 B3DK94 A0A1B6CK41 A0A1W4YLQ0 A0A1W4YW35 A0A1X7UFI5 A0A3B4BGA9 Q5RHH4
Pubmed
EMBL
BABH01011905
RSAL01000137
RVE46188.1
NWSH01000021
PCG80678.1
ODYU01006762
+ More
SOQ48901.1 KQ459606 KPI91679.1 CH477327 EAT43464.1 CH477329 EAT43404.1 AE014296 CH480817 EDW50172.1 CM002912 KMY97056.1 KMY97058.1 KMY97057.1 DS231817 EDS39627.1 CH954178 EDV50276.1 CH940647 EDW69772.1 OUUW01000012 SPP87638.1 CM000159 EDW92960.1 AXCN02000907 CH902618 EDV38716.1 CH964062 EDW78746.1 CH933809 EDW18313.1 CCAG010018360 JXJN01025783 JXJN01013485 AAAB01008960 EAL39797.1 APCN01004435 ATLV01004460 KE524214 KFB35084.1 CH379069 EAL29770.2 CH916366 EDV96827.1 KRT08360.1 AXCM01002609 JH431432 JRES01000493 KNC30792.1 CVRI01000042 CRK95655.1 DS497723 EFA12908.1 ADMH02001057 ETN64252.1 GEFM01003764 JAP72032.1 GEGO01003495 JAR91909.1 KK852721 KDR17746.1 JYDP01000232 KRZ02367.1 JYDO01000004 KRZ80046.1 HACA01012180 CDW29541.1 KRZ80045.1 KRZ80047.1 DS235281 EEB14362.1 ABJB010099173 ABJB010179507 ABJB010422567 ABJB010475508 ABJB010600645 ABJB010696855 ABJB010818241 ABJB010917940 ABJB011057937 ABJB011105378 DS886054 EEC15263.1 KRZ02369.1 JYDJ01000189 KRX40774.1 KRZ02366.1 KRZ02365.1 KRZ02368.1 JYDQ01000001 KRY23782.1 KRY23781.1 APGK01055682 APGK01055683 KB741269 ENN71435.1 JYDK01000012 KRX83551.1 JYDM01000089 KRZ88148.1 JYDW01000003 KRZ63058.1 ABLF02025628 ABLF02025629 ABLF02025630 ABLF02025631 GFTR01008877 JAW07549.1 CP012525 ALC42884.1 JYDH01000008 KRY41446.1 JYDI01000062 KRY54929.1 KRY54928.1 JT406107 AHH37456.1 KB632330 ERL92593.1 GGMS01010883 MBY80086.1 GEDC01002054 JAS35244.1 GEDC01018223 JAS19075.1 NEDP02005561 OWF38575.1 BC163767 AAI63767.1 GEDC01023434 JAS13864.1 AY618923 BX511178
SOQ48901.1 KQ459606 KPI91679.1 CH477327 EAT43464.1 CH477329 EAT43404.1 AE014296 CH480817 EDW50172.1 CM002912 KMY97056.1 KMY97058.1 KMY97057.1 DS231817 EDS39627.1 CH954178 EDV50276.1 CH940647 EDW69772.1 OUUW01000012 SPP87638.1 CM000159 EDW92960.1 AXCN02000907 CH902618 EDV38716.1 CH964062 EDW78746.1 CH933809 EDW18313.1 CCAG010018360 JXJN01025783 JXJN01013485 AAAB01008960 EAL39797.1 APCN01004435 ATLV01004460 KE524214 KFB35084.1 CH379069 EAL29770.2 CH916366 EDV96827.1 KRT08360.1 AXCM01002609 JH431432 JRES01000493 KNC30792.1 CVRI01000042 CRK95655.1 DS497723 EFA12908.1 ADMH02001057 ETN64252.1 GEFM01003764 JAP72032.1 GEGO01003495 JAR91909.1 KK852721 KDR17746.1 JYDP01000232 KRZ02367.1 JYDO01000004 KRZ80046.1 HACA01012180 CDW29541.1 KRZ80045.1 KRZ80047.1 DS235281 EEB14362.1 ABJB010099173 ABJB010179507 ABJB010422567 ABJB010475508 ABJB010600645 ABJB010696855 ABJB010818241 ABJB010917940 ABJB011057937 ABJB011105378 DS886054 EEC15263.1 KRZ02369.1 JYDJ01000189 KRX40774.1 KRZ02366.1 KRZ02365.1 KRZ02368.1 JYDQ01000001 KRY23782.1 KRY23781.1 APGK01055682 APGK01055683 KB741269 ENN71435.1 JYDK01000012 KRX83551.1 JYDM01000089 KRZ88148.1 JYDW01000003 KRZ63058.1 ABLF02025628 ABLF02025629 ABLF02025630 ABLF02025631 GFTR01008877 JAW07549.1 CP012525 ALC42884.1 JYDH01000008 KRY41446.1 JYDI01000062 KRY54929.1 KRY54928.1 JT406107 AHH37456.1 KB632330 ERL92593.1 GGMS01010883 MBY80086.1 GEDC01002054 JAS35244.1 GEDC01018223 JAS19075.1 NEDP02005561 OWF38575.1 BC163767 AAI63767.1 GEDC01023434 JAS13864.1 AY618923 BX511178
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000008820
UP000192221
+ More
UP000000803 UP000001292 UP000002320 UP000008711 UP000008792 UP000075885 UP000268350 UP000002282 UP000075886 UP000075881 UP000007801 UP000007798 UP000092445 UP000009192 UP000092444 UP000092460 UP000078200 UP000076407 UP000075902 UP000007062 UP000075900 UP000075840 UP000075903 UP000030765 UP000001819 UP000001070 UP000076408 UP000075920 UP000075883 UP000075884 UP000091820 UP000095301 UP000069272 UP000037069 UP000183832 UP000007266 UP000095300 UP000000673 UP000027135 UP000055024 UP000054843 UP000009046 UP000001555 UP000055048 UP000054783 UP000019118 UP000054673 UP000054924 UP000054721 UP000007819 UP000092553 UP000046395 UP000054776 UP000054653 UP000221080 UP000030742 UP000261440 UP000257160 UP000265080 UP000242188 UP000192224 UP000007879 UP000261520 UP000000437
UP000000803 UP000001292 UP000002320 UP000008711 UP000008792 UP000075885 UP000268350 UP000002282 UP000075886 UP000075881 UP000007801 UP000007798 UP000092445 UP000009192 UP000092444 UP000092460 UP000078200 UP000076407 UP000075902 UP000007062 UP000075900 UP000075840 UP000075903 UP000030765 UP000001819 UP000001070 UP000076408 UP000075920 UP000075883 UP000075884 UP000091820 UP000095301 UP000069272 UP000037069 UP000183832 UP000007266 UP000095300 UP000000673 UP000027135 UP000055024 UP000054843 UP000009046 UP000001555 UP000055048 UP000054783 UP000019118 UP000054673 UP000054924 UP000054721 UP000007819 UP000092553 UP000046395 UP000054776 UP000054653 UP000221080 UP000030742 UP000261440 UP000257160 UP000265080 UP000242188 UP000192224 UP000007879 UP000261520 UP000000437
Pfam
Interpro
IPR011990
TPR-like_helical_dom_sf
+ More
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR024977 Apc4_WD40_dom
IPR017986 WD40_repeat_dom
IPR017956 AT_hook_DNA-bd_motif
IPR038489 SUFU_C_sf
IPR018391 PQQ_beta_propeller_repeat
IPR024314 SUFU_C
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
IPR020941 SUFU-like_domain
IPR011009 Kinase-like_dom_sf
IPR037181 SUFU_N
IPR008271 Ser/Thr_kinase_AS
IPR002372 PQQ_repeat
IPR017441 Protein_kinase_ATP_BS
IPR019734 TPR_repeat
IPR007714 CFA20_dom
IPR036396 Cyt_P450_sf
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR002401 Cyt_P450_E_grp-I
IPR011044 Quino_amine_DH_bsu
IPR006692 Coatomer_WD-assoc_reg
IPR016024 ARM-type_fold
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR024977 Apc4_WD40_dom
IPR017986 WD40_repeat_dom
IPR017956 AT_hook_DNA-bd_motif
IPR038489 SUFU_C_sf
IPR018391 PQQ_beta_propeller_repeat
IPR024314 SUFU_C
IPR011047 Quinoprotein_ADH-like_supfam
IPR000719 Prot_kinase_dom
IPR020941 SUFU-like_domain
IPR011009 Kinase-like_dom_sf
IPR037181 SUFU_N
IPR008271 Ser/Thr_kinase_AS
IPR002372 PQQ_repeat
IPR017441 Protein_kinase_ATP_BS
IPR019734 TPR_repeat
IPR007714 CFA20_dom
IPR036396 Cyt_P450_sf
IPR017972 Cyt_P450_CS
IPR001128 Cyt_P450
IPR002401 Cyt_P450_E_grp-I
IPR011044 Quino_amine_DH_bsu
IPR006692 Coatomer_WD-assoc_reg
IPR016024 ARM-type_fold
SUPFAM
ProteinModelPortal
H9JQE4
A0A3S2NAW4
A0A2A4K9V4
A0A2H1W743
A0A194PG50
A0A1S4F9Q5
+ More
Q17B06 A0A1W4W4F3 Q17AW7 Q9W040 B4HW57 A0A0J9RMB7 A0A0J9RMU1 B0W088 B3NBF4 B4LBX2 A0A182PGF5 A0A3B0KUL4 B4PEC2 A0A182QPT3 A0A182JUZ2 B3M755 B4N307 A0A1A9Z4J4 B4KV02 A0A1B0F9U2 A0A1B0AKI5 A0A1A9VEC2 A0A1B0BFI1 A0A182XD40 A0A182UJV6 Q5TRR4 A0A182RX30 A0A182HMH4 A0A182VL23 A0A084VAU0 Q2LYU0 B4J0J1 A0A182YPZ2 A0A0R3P5G5 A0A182WAJ7 A0A182MI61 A0A182N7Y3 T1ISG8 A0A1A9W7J7 A0A1I8NB59 A0A182FHA6 A0A0L0CF00 A0A1J1I5Q0 D7EJZ7 A0A1I8PPJ7 W5JIP2 A0A131Y0L6 A0A147BMA5 A0A067RFG0 A0A0V1GW14 A0A0V1N8M8 A0A0K2TVY2 A0A0V1N7V3 A0A0V1N841 E0VLV6 B7Q8U1 A0A0V1GVW5 A0A0V0TP49 A0A0V1GVJ3 A0A0V1GW08 A0A0V1GVV6 A0A0V1AG17 A0A0V1AG32 N6TQB8 A0A0V0X6A3 A0A0V1NVW4 A0A0V1LV56 J9L180 A0A224XHN2 A0A0M4EC07 A0A0N5E211 A0A0V1BWV4 A0A0V1D0D9 A0A0V1D0C5 W5U640 U4UJF4 A0A2S2QQN7 A0A3B4BQU6 A0A3B4BQG8 A0A3B4BLU9 A0A3Q1AR83 A0A3P8SZX4 A0A1B6EBJ3 A0A1B6D0F4 A0A210PPZ5 B3DK94 A0A1B6CK41 A0A1W4YLQ0 A0A1W4YW35 A0A1X7UFI5 A0A3B4BGA9 Q5RHH4
Q17B06 A0A1W4W4F3 Q17AW7 Q9W040 B4HW57 A0A0J9RMB7 A0A0J9RMU1 B0W088 B3NBF4 B4LBX2 A0A182PGF5 A0A3B0KUL4 B4PEC2 A0A182QPT3 A0A182JUZ2 B3M755 B4N307 A0A1A9Z4J4 B4KV02 A0A1B0F9U2 A0A1B0AKI5 A0A1A9VEC2 A0A1B0BFI1 A0A182XD40 A0A182UJV6 Q5TRR4 A0A182RX30 A0A182HMH4 A0A182VL23 A0A084VAU0 Q2LYU0 B4J0J1 A0A182YPZ2 A0A0R3P5G5 A0A182WAJ7 A0A182MI61 A0A182N7Y3 T1ISG8 A0A1A9W7J7 A0A1I8NB59 A0A182FHA6 A0A0L0CF00 A0A1J1I5Q0 D7EJZ7 A0A1I8PPJ7 W5JIP2 A0A131Y0L6 A0A147BMA5 A0A067RFG0 A0A0V1GW14 A0A0V1N8M8 A0A0K2TVY2 A0A0V1N7V3 A0A0V1N841 E0VLV6 B7Q8U1 A0A0V1GVW5 A0A0V0TP49 A0A0V1GVJ3 A0A0V1GW08 A0A0V1GVV6 A0A0V1AG17 A0A0V1AG32 N6TQB8 A0A0V0X6A3 A0A0V1NVW4 A0A0V1LV56 J9L180 A0A224XHN2 A0A0M4EC07 A0A0N5E211 A0A0V1BWV4 A0A0V1D0D9 A0A0V1D0C5 W5U640 U4UJF4 A0A2S2QQN7 A0A3B4BQU6 A0A3B4BQG8 A0A3B4BLU9 A0A3Q1AR83 A0A3P8SZX4 A0A1B6EBJ3 A0A1B6D0F4 A0A210PPZ5 B3DK94 A0A1B6CK41 A0A1W4YLQ0 A0A1W4YW35 A0A1X7UFI5 A0A3B4BGA9 Q5RHH4
Ontologies
GO
GO:0031514
GO:1905515
GO:0005929
GO:0007018
GO:0060271
GO:0030990
GO:0042073
GO:0005871
GO:0007275
GO:0097730
GO:0005930
GO:0036064
GO:0030992
GO:0005524
GO:0003677
GO:0004672
GO:0020037
GO:0005506
GO:0016705
GO:0006886
GO:0030117
GO:0016192
GO:0005198
GO:0048793
GO:0032006
GO:0060041
GO:0045494
GO:0016021
GO:0005515
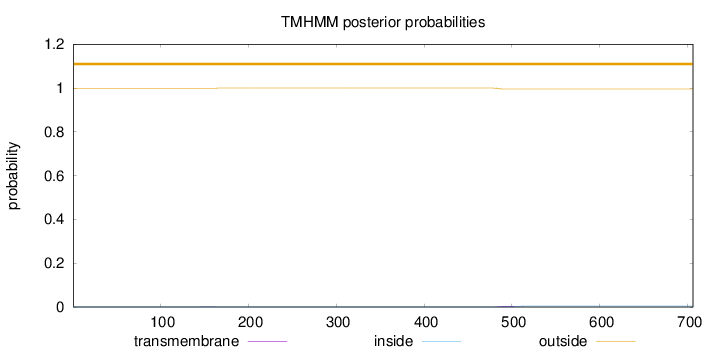
Topology
Subcellular location
Length:
706
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08984
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00045
outside
1 - 706
Population Genetic Test Statistics
Pi
183.542251
Theta
164.536488
Tajima's D
0.229733
CLR
0.265062
CSRT
0.433978301084946
Interpretation
Uncertain
