Gene
KWMTBOMO06708
Pre Gene Modal
BGIBMGA011735
Annotation
PREDICTED:_ecto-NOX_disulfide-thiol_exchanger_2_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.38
Sequence
CDS
ATGCAAGGACATCGAAGGCATAGGTCTAGATCGCCTTTGAAAGAAAGCAATGGTGGCCGCCGAAGAGATGGAAACAGCCATGAAAGCAAGAGGGCTAGCGATTTGCCTAATGCAAATGTCATGAACGCAATGATGTTACAGAATATGTATCAACAAAGTGGCATGATGATGGCTGGTGGAATGTATCCAAATATGATTATGCCTAGTTCTGTGATGGGTGGAGGAATAATGCCATCAACTGGCATGGAGATAATGTCAACCACAGGAATAGAAATAATGCCCCAGCCTTCTATGGATGTGTCTGCAATAGGGACTCCAACAATATCCACACCAGGTAGTTCAACAATTGATATGAATATGATGGGAGGAATGGTGGTAGATCCTGCAATGATGGGAATTTATGGAATGACTGGAGACCTATCAAACTTGCAAGAAAAGAAGGAAATTGTGTTCAAAAATTGTAAGCTAATTCCTCCAGATTCTGGTACTCCTCAACCTCCTCGTCGTGCTAAACCACCTGGTTGTCGTACTATTTTTATTGGAGGCCTCCCAGATAAAATACGTGAAAGTTCTGTCAGAGAGATATTTGAAAACTATGGCAGGATTATATCATTAAGATTATCTAGAAAAAATTTCTGTCACATACGGTATGACAGAGAATCCTGTGTTGATGCGGCGATGACAATTTCGGGTTATCGAGTGAAACTTATAAATAAAGATGGAGACAATAATGGAGAAGAACAGGATACCCATGGTACAGTGGGTTGGTTACATGTTGACTATGCCATGAGCAGAGATGATCAAAATGAATTTGAAAGACGACAAAGACAAGCATTACGAGTTCAGCAAGCTCAAATGCAGCAGTTGAATGCTCAACAAGAAATTTTAAACTCCACAAATGAATATAGGCAATCACCATCTCCAGTAAGGATCCAGCCATATTCAAACCCTGCTATGGTACAACTCTCTGAAAAAATTAAAAGTGAAGAGCATTTTTCAAGTGCCCTACCAACCTTACTTGCATGGCTTGAACGTGGTGAATGCTCTAAAAAAAATTCAAATCAGTTTTACTCCATGATTCAGGCTACGAATTCTCATATACGTAGATTATTTAATGAAAAAATGCAGGCTGAAGAAGAGCTGCAGGAGTGTAAGGACAGAATTAGGAAAACAATGGAAAATGTGATTGATCAACTCGAACAGGTGGCGAAAGTGTTTACAGCATCTACCCACCAACGAGTCTGGGATCATTTCACCAAACCCCAACGAAAGAACATTGAGACATGGCAGAAAATGGCACAGGAATTTAGTACACTGAAAGAAGAATTGTTTGAGAAATTTTACAATGACGACGACTATAATGGCTGTAAGGCATCGGATGGGTACACTGAAGACATTCAAGTTTTAAAGCGCGAAAATGAAAGTCTGCGGTTTCAATTGGAAGCTTACAAGAATGAAGTTGATTTAATAAAAGCAGACTCTCAAAAAGAGATGGAGAAATTCAAAGCGCAGTTCATTGCTCGTCAAGCTTTGCAAGGAATTTTTGAAAACAGGAACGTAAGTGAGCCTCCCCTACCATCTCCGGTAACGAAGCCTCCGCCTCCGCCACCGTTGCCTGATGATATGAATTCGAAAGTTCATTTGAACGAGACAGTCGAAGTCGGCTGTGGAGAAGCCAAATTAATTGGTGTAATGTCAGCGTTTTTGCAGGTGCACCCGCAAGGAGCGAGCCTGGACTACGTGTTGTCGTACGTGCGCGCCGTGTTCCCGGCCGTGTCGCAGGCGGCCGTGCACCGCGTGCTCCAGAGACACGCGGACGTGTTCAGCCGCACCACGTCAGGGGTCGGAGCGAACATCGAGCACCGCTGGAACTTTGTTGCCTTCCAGAACGCCGACTCTGGATCATAG
Protein
MQGHRRHRSRSPLKESNGGRRRDGNSHESKRASDLPNANVMNAMMLQNMYQQSGMMMAGGMYPNMIMPSSVMGGGIMPSTGMEIMSTTGIEIMPQPSMDVSAIGTPTISTPGSSTIDMNMMGGMVVDPAMMGIYGMTGDLSNLQEKKEIVFKNCKLIPPDSGTPQPPRRAKPPGCRTIFIGGLPDKIRESSVREIFENYGRIISLRLSRKNFCHIRYDRESCVDAAMTISGYRVKLINKDGDNNGEEQDTHGTVGWLHVDYAMSRDDQNEFERRQRQALRVQQAQMQQLNAQQEILNSTNEYRQSPSPVRIQPYSNPAMVQLSEKIKSEEHFSSALPTLLAWLERGECSKKNSNQFYSMIQATNSHIRRLFNEKMQAEEELQECKDRIRKTMENVIDQLEQVAKVFTASTHQRVWDHFTKPQRKNIETWQKMAQEFSTLKEELFEKFYNDDDYNGCKASDGYTEDIQVLKRENESLRFQLEAYKNEVDLIKADSQKEMEKFKAQFIARQALQGIFENRNVSEPPLPSPVTKPPPPPPLPDDMNSKVHLNETVEVGCGEAKLIGVMSAFLQVHPQGASLDYVLSYVRAVFPAVSQAAVHRVLQRHADVFSRTTSGVGANIEHRWNFVAFQNADSGS
Summary
Uniprot
H9JQC9
A0A2A4K9E2
A0A0N1IEL9
A0A1E1W917
A0A212FGU7
A0A2H1WUJ1
+ More
A0A0L7LRL1 A0A2A4JK90 A0A0L7QSN1 A0A1Y1N132 A0A1S3HU38 A0A088A3I5 D6WDD0 K7IQY5 A0A0C9PRB5 A0A232F3L4 T1HM91 A0A1S3K543 E0VIU7 A0A1B6DUN6 A0A023F1J3 A0A026WUA5 A0A3L8DFV8 A0A1B6CWL8 A0A154NYH3 F4WYV3 A0A151JBT3 A0A195FAX4 A0A158NC63 A0A195CMI7 A0A151XCR1 A0A1B6KCA5 A0A151HZW9 A0A1B6KP47 A0A1Z5LDL2 E9IM10 A0A3P8URZ4 A0A3B4FPQ7 A0A3P8NGD5 A0A3Q4H0T4 A0A0K8S492 N6SUS1 A0A2Y9NCF5 I3J2F3 A0A2U4BFV6 A0A3P9BNA5 A0A341CBT3 A0A293LRE3 V4AL77 A0A3Q3SQI3 A0A3Q0DPX4 A0A3P9A8Q3 A0A3Q2VWY2 A0A3Q4H0R3 A0A3B4FPU3 A0A3Q2PQV6 A0A3Q4HGD6 A0A3B4FR33 A0A3P8NG84 A0A3P9BN93 M3ZDF0 A0A3P8NG78 A0A3P9BMA6 A0A3Q3GT58 A0A3B5MMR6 A0A3Q2YT12 A0A3Q2VVW7 A0A3B4FPK4 A0A3Q2VXK5 A0A3Q3D8R1 A0A3Q2VWX4 A0A3Q0S805 A0A3P8NG69 A0A3P9BN55 A0A3Q2YT20 A0A1A8QCK4 A0A1A8NGK2 A0A087X844 A0A1A7ZQD4 A0A1A8NUV4 A0A3Q3MML1 A0A3Q3MMJ2 A0A3P9A8R2 A0A3B5Q624 E2B5B6 A0A3B5PZ03 J9K198 A0A3Q2CDS8 L7MEF9 A0A3B4BBB1 A0A3P8S2J7 A0A3Q1BH22 A0A3Q1GMM1 W5KWF0
A0A0L7LRL1 A0A2A4JK90 A0A0L7QSN1 A0A1Y1N132 A0A1S3HU38 A0A088A3I5 D6WDD0 K7IQY5 A0A0C9PRB5 A0A232F3L4 T1HM91 A0A1S3K543 E0VIU7 A0A1B6DUN6 A0A023F1J3 A0A026WUA5 A0A3L8DFV8 A0A1B6CWL8 A0A154NYH3 F4WYV3 A0A151JBT3 A0A195FAX4 A0A158NC63 A0A195CMI7 A0A151XCR1 A0A1B6KCA5 A0A151HZW9 A0A1B6KP47 A0A1Z5LDL2 E9IM10 A0A3P8URZ4 A0A3B4FPQ7 A0A3P8NGD5 A0A3Q4H0T4 A0A0K8S492 N6SUS1 A0A2Y9NCF5 I3J2F3 A0A2U4BFV6 A0A3P9BNA5 A0A341CBT3 A0A293LRE3 V4AL77 A0A3Q3SQI3 A0A3Q0DPX4 A0A3P9A8Q3 A0A3Q2VWY2 A0A3Q4H0R3 A0A3B4FPU3 A0A3Q2PQV6 A0A3Q4HGD6 A0A3B4FR33 A0A3P8NG84 A0A3P9BN93 M3ZDF0 A0A3P8NG78 A0A3P9BMA6 A0A3Q3GT58 A0A3B5MMR6 A0A3Q2YT12 A0A3Q2VVW7 A0A3B4FPK4 A0A3Q2VXK5 A0A3Q3D8R1 A0A3Q2VWX4 A0A3Q0S805 A0A3P8NG69 A0A3P9BN55 A0A3Q2YT20 A0A1A8QCK4 A0A1A8NGK2 A0A087X844 A0A1A7ZQD4 A0A1A8NUV4 A0A3Q3MML1 A0A3Q3MMJ2 A0A3P9A8R2 A0A3B5Q624 E2B5B6 A0A3B5PZ03 J9K198 A0A3Q2CDS8 L7MEF9 A0A3B4BBB1 A0A3P8S2J7 A0A3Q1BH22 A0A3Q1GMM1 W5KWF0
Pubmed
EMBL
BABH01011926
NWSH01000021
PCG80689.1
KQ460499
KPJ14307.1
GDQN01007591
+ More
JAT83463.1 AGBW02008593 OWR52966.1 ODYU01011154 SOQ56720.1 JTDY01000241 KOB78100.1 NWSH01001237 PCG72014.1 KQ414758 KOC61476.1 GEZM01016808 JAV91138.1 KQ971321 EEZ99476.1 GBYB01003783 JAG73550.1 NNAY01001025 OXU25436.1 ACPB03012516 ACPB03012517 DS235206 EEB13303.1 GEDC01007943 JAS29355.1 GBBI01003848 JAC14864.1 KK107109 EZA59241.1 QOIP01000008 RLU19181.1 GEDC01019392 JAS17906.1 KQ434783 KZC04719.1 GL888463 EGI60607.1 KQ979120 KYN22576.1 KQ981727 KYN37184.1 ADTU01011667 ADTU01011668 KQ977565 KYN01941.1 KQ982298 KYQ58162.1 GEBQ01030911 JAT09066.1 KQ976642 KYM78660.1 GEBQ01026771 JAT13206.1 GFJQ02001491 JAW05479.1 GL764129 EFZ18537.1 GBRD01017843 GBRD01017840 JAG47984.1 APGK01055685 APGK01055686 KB741269 KB632330 ENN71439.1 ERL92588.1 AERX01013135 GFWV01005648 MAA30378.1 KB199651 ESP04949.1 HAEI01004911 SBR90904.1 HAEF01000833 HAEG01002913 SBR68195.1 AYCK01014407 AYCK01014408 HADY01006479 HAEJ01013665 SBP44964.1 HAEH01004024 SBR72831.1 GL445814 EFN89111.1 ABLF02020813 ABLF02020814 GACK01003396 JAA61638.1
JAT83463.1 AGBW02008593 OWR52966.1 ODYU01011154 SOQ56720.1 JTDY01000241 KOB78100.1 NWSH01001237 PCG72014.1 KQ414758 KOC61476.1 GEZM01016808 JAV91138.1 KQ971321 EEZ99476.1 GBYB01003783 JAG73550.1 NNAY01001025 OXU25436.1 ACPB03012516 ACPB03012517 DS235206 EEB13303.1 GEDC01007943 JAS29355.1 GBBI01003848 JAC14864.1 KK107109 EZA59241.1 QOIP01000008 RLU19181.1 GEDC01019392 JAS17906.1 KQ434783 KZC04719.1 GL888463 EGI60607.1 KQ979120 KYN22576.1 KQ981727 KYN37184.1 ADTU01011667 ADTU01011668 KQ977565 KYN01941.1 KQ982298 KYQ58162.1 GEBQ01030911 JAT09066.1 KQ976642 KYM78660.1 GEBQ01026771 JAT13206.1 GFJQ02001491 JAW05479.1 GL764129 EFZ18537.1 GBRD01017843 GBRD01017840 JAG47984.1 APGK01055685 APGK01055686 KB741269 KB632330 ENN71439.1 ERL92588.1 AERX01013135 GFWV01005648 MAA30378.1 KB199651 ESP04949.1 HAEI01004911 SBR90904.1 HAEF01000833 HAEG01002913 SBR68195.1 AYCK01014407 AYCK01014408 HADY01006479 HAEJ01013665 SBP44964.1 HAEH01004024 SBR72831.1 GL445814 EFN89111.1 ABLF02020813 ABLF02020814 GACK01003396 JAA61638.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000037510
UP000053825
+ More
UP000085678 UP000005203 UP000007266 UP000002358 UP000215335 UP000015103 UP000009046 UP000053097 UP000279307 UP000076502 UP000007755 UP000078492 UP000078541 UP000005205 UP000078542 UP000075809 UP000078540 UP000265120 UP000261460 UP000265100 UP000261580 UP000019118 UP000030742 UP000248483 UP000005207 UP000245320 UP000265160 UP000252040 UP000030746 UP000261640 UP000189704 UP000265140 UP000264840 UP000265000 UP000002852 UP000264800 UP000261380 UP000264820 UP000261340 UP000028760 UP000008237 UP000007819 UP000265020 UP000261520 UP000265080 UP000257160 UP000257200 UP000018467
UP000085678 UP000005203 UP000007266 UP000002358 UP000215335 UP000015103 UP000009046 UP000053097 UP000279307 UP000076502 UP000007755 UP000078492 UP000078541 UP000005205 UP000078542 UP000075809 UP000078540 UP000265120 UP000261460 UP000265100 UP000261580 UP000019118 UP000030742 UP000248483 UP000005207 UP000245320 UP000265160 UP000252040 UP000030746 UP000261640 UP000189704 UP000265140 UP000264840 UP000265000 UP000002852 UP000264800 UP000261380 UP000264820 UP000261340 UP000028760 UP000008237 UP000007819 UP000265020 UP000261520 UP000265080 UP000257160 UP000257200 UP000018467
PRIDE
Pfam
PF00076 RRM_1
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
CDD
ProteinModelPortal
H9JQC9
A0A2A4K9E2
A0A0N1IEL9
A0A1E1W917
A0A212FGU7
A0A2H1WUJ1
+ More
A0A0L7LRL1 A0A2A4JK90 A0A0L7QSN1 A0A1Y1N132 A0A1S3HU38 A0A088A3I5 D6WDD0 K7IQY5 A0A0C9PRB5 A0A232F3L4 T1HM91 A0A1S3K543 E0VIU7 A0A1B6DUN6 A0A023F1J3 A0A026WUA5 A0A3L8DFV8 A0A1B6CWL8 A0A154NYH3 F4WYV3 A0A151JBT3 A0A195FAX4 A0A158NC63 A0A195CMI7 A0A151XCR1 A0A1B6KCA5 A0A151HZW9 A0A1B6KP47 A0A1Z5LDL2 E9IM10 A0A3P8URZ4 A0A3B4FPQ7 A0A3P8NGD5 A0A3Q4H0T4 A0A0K8S492 N6SUS1 A0A2Y9NCF5 I3J2F3 A0A2U4BFV6 A0A3P9BNA5 A0A341CBT3 A0A293LRE3 V4AL77 A0A3Q3SQI3 A0A3Q0DPX4 A0A3P9A8Q3 A0A3Q2VWY2 A0A3Q4H0R3 A0A3B4FPU3 A0A3Q2PQV6 A0A3Q4HGD6 A0A3B4FR33 A0A3P8NG84 A0A3P9BN93 M3ZDF0 A0A3P8NG78 A0A3P9BMA6 A0A3Q3GT58 A0A3B5MMR6 A0A3Q2YT12 A0A3Q2VVW7 A0A3B4FPK4 A0A3Q2VXK5 A0A3Q3D8R1 A0A3Q2VWX4 A0A3Q0S805 A0A3P8NG69 A0A3P9BN55 A0A3Q2YT20 A0A1A8QCK4 A0A1A8NGK2 A0A087X844 A0A1A7ZQD4 A0A1A8NUV4 A0A3Q3MML1 A0A3Q3MMJ2 A0A3P9A8R2 A0A3B5Q624 E2B5B6 A0A3B5PZ03 J9K198 A0A3Q2CDS8 L7MEF9 A0A3B4BBB1 A0A3P8S2J7 A0A3Q1BH22 A0A3Q1GMM1 W5KWF0
A0A0L7LRL1 A0A2A4JK90 A0A0L7QSN1 A0A1Y1N132 A0A1S3HU38 A0A088A3I5 D6WDD0 K7IQY5 A0A0C9PRB5 A0A232F3L4 T1HM91 A0A1S3K543 E0VIU7 A0A1B6DUN6 A0A023F1J3 A0A026WUA5 A0A3L8DFV8 A0A1B6CWL8 A0A154NYH3 F4WYV3 A0A151JBT3 A0A195FAX4 A0A158NC63 A0A195CMI7 A0A151XCR1 A0A1B6KCA5 A0A151HZW9 A0A1B6KP47 A0A1Z5LDL2 E9IM10 A0A3P8URZ4 A0A3B4FPQ7 A0A3P8NGD5 A0A3Q4H0T4 A0A0K8S492 N6SUS1 A0A2Y9NCF5 I3J2F3 A0A2U4BFV6 A0A3P9BNA5 A0A341CBT3 A0A293LRE3 V4AL77 A0A3Q3SQI3 A0A3Q0DPX4 A0A3P9A8Q3 A0A3Q2VWY2 A0A3Q4H0R3 A0A3B4FPU3 A0A3Q2PQV6 A0A3Q4HGD6 A0A3B4FR33 A0A3P8NG84 A0A3P9BN93 M3ZDF0 A0A3P8NG78 A0A3P9BMA6 A0A3Q3GT58 A0A3B5MMR6 A0A3Q2YT12 A0A3Q2VVW7 A0A3B4FPK4 A0A3Q2VXK5 A0A3Q3D8R1 A0A3Q2VWX4 A0A3Q0S805 A0A3P8NG69 A0A3P9BN55 A0A3Q2YT20 A0A1A8QCK4 A0A1A8NGK2 A0A087X844 A0A1A7ZQD4 A0A1A8NUV4 A0A3Q3MML1 A0A3Q3MMJ2 A0A3P9A8R2 A0A3B5Q624 E2B5B6 A0A3B5PZ03 J9K198 A0A3Q2CDS8 L7MEF9 A0A3B4BBB1 A0A3P8S2J7 A0A3Q1BH22 A0A3Q1GMM1 W5KWF0
PDB
5CA5
E-value=0.00675604,
Score=95
Ontologies
GO
PANTHER
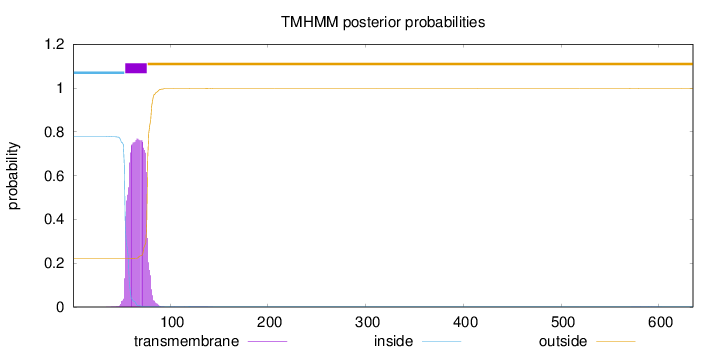
Topology
Length:
635
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.34386
Exp number, first 60 AAs:
4.32383
Total prob of N-in:
0.77943
inside
1 - 53
TMhelix
54 - 76
outside
77 - 635
Population Genetic Test Statistics
Pi
189.355625
Theta
168.248463
Tajima's D
-0.940477
CLR
58.017932
CSRT
0.149242537873106
Interpretation
Uncertain
