Gene
KWMTBOMO06705 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011754
Annotation
PREDICTED:_ATP-dependent_RNA_helicase_dbp2-like_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.851
Sequence
CDS
ATGCAGTACAACGGGTTTCACAATCAACATCCGATTTTCAGGCCTCAGCCAGACCGCAATGAACAATTCGGTAAAATTCCATACATTAAAGGTCTTCCTGATTTTGGAGGACCTAAAAACTTTGGACCAAGACCTAATATGATGAACAAAAACTTCAGACCTCGAAATGATTTTAATGAAGTAAAGAATGATTATAATACTAAAAATGATGGTAATCAGAATGATTTTGGTGGTCCAAAACAATTTAGACCCAGAAATAATTTTAACAATGGTAACCAACCACCAAAGAAAAACAACTTTAATGGAGACAAATCACCAGGAAATATGCAATATGGCAATAAAAATGATTTTGGCGGTCCTAAACAACAAAATTATAACAAGAATTATGGGCCTAAAACATACAACAACCAGAATTGCTATGGGAATGAGCAGCCTGAATTTATTCCTAGGCAATCTTACAGTCCAAACTCTGTTCAACATTCTTTGAATGATCGCAAACTACAGAGCAAAAAAGCAAAACATCCTGGTGATAGCTTGTTGAAGCCAATGTGGGATCTAACTAAATTAGAACCTATTCAGAAAGACTTTTACAAGCCTCATGTTAATGTAGATGCTCGCTCTGAGGACGATGTGCAAAGATTCCGTGTTACAAAAGAGATTACAGTTAGTGGCAATAATATCCCACGTCCTAACCAATTATTTGAGGAAGGAAACTTCCCCGACCACATTATGAGCACTATTAAGGAACAGCCAAATTTGATAGAGCCCACTGGTATTCAAGCTCAAGGTTGGCCAATAGCTTTATCAGGACGTGACATGGTTGGTATTGCCTCTACTGGTTCAGGAAAAACTTTAGCTTACATGTTGCCTGCGGCTGTGCATATTGTTCATCAACAGCGCATTCAGCGAGGAGATGGGCCAATTGCGCTTATTTTAGCACCTACTCGAGAACTGGCACAGCAAATCCAGTCAGTAGCCCAGGCTTACAGTGCTCGTGGTTGCATAAGAAACACATGCCTTTTTGGAGGTTCACCAAAAGGACCACAAGCCAGAGATTTAGAGCGAGGTGTTGAAATTGTTATTGCCACACCTGGAAGGTTAATTGATTTCCTTGAACGAGGCACTACTAACTTAAGACGCTGCACATATTTAGTTCTGGATGAAGCAGATAGAATGCTTGACATGGGATTTGAACCTCAAATCCGTAAGATAATTGAACAGATTCGCCCTGACCGTCAAGTACTGATGTGGTCTGCCACATGGCCTAAGGAAATACAGGCTTTGGCTGAAGATTTCTTGACCGATTACATCAAAGTTAATATTGGCTCCCTAAACTTGTCTGCCAACAACAACATCAAACAGATTATTGAGATTTGTGAAGAACATGAGAAGGAAGTAAAATTAACTAATTTATTAAAAGAAATTGCTTCTGAAAAAGAAAATAAAGTAATTGTATTTGTAGAGACAAAGAAAAAGGTGGATGATATTGCACGTGCAGTTCGACGCGGTGGATATAAGGCACTTGCTATTCATGGTGACAAATCCCAACCTGAACGTGATGCTGTTCTTACAGAATTCAGAAATGGTTCAACAACCATTCTGATTGCTACTGATGTCGCAGCTCGTGGTCTTGATGTTGAAGATGTCAAATTTGTTGTAAATTTTGACTACCCTAACTCTTCGGAAGACTATATTCATAGAATTGGTCGTACTGGACGGTGCCAACAATCTGGAACTGCATATACATACTTCACGAGTGGTGATTGCCGCCAAGCTCGAGGTCTGCTTGCAGTACTTCGGGAAACTGGACAAAATCCTCCAACTAAATTAGCTGATATGGCTAGAAATAATAACAATAATTCAGGTCGGAATCGCTGGCAACAACGCAAGGATAAAAATAGTGGATCCAACTCACCACAGCAAATGAGCAACCAATGGAACAACAAAATGGCCAATGGCAGCATTGAAGATGGTCAAAACATGTACCAAAATCGTAGCATGAACCAACAGGCACCACGATTTTCTAACCAGAACCAAGGAATTCAGAGTTACAGACAAAACAACTATCAGAAACAAGTACCTTTCCCAAGCCCCAATGTTTTCGAGTCCATGGGAAATTATCAAGGTGGATATAACAATGGTTACCAAAATACTTACAACCAAAATGGATATGGTCAACGGCCCTTCAACAACCAGCGTAATAATGGGCAACAATATCGTAACAATGGTACAAATGGCAACAAAGCCAGTGGCTCTGGTTCTTCATTTGGTTCCCCACCTCCACATTTTGCAACACCTCCTCATTACGCACAGGTGCATCCACATCATCAGGATATGCTTGGTGGAAAATTTTACGGTAGTGGTGTCGGTGGGGGCGGACCCTGTGGATATCAAGCTGCCGTGGGCGCTGGTGTTCCCTACCCGCACTTGCCCCCAGGGCCACTACTATATGCAGCCCCTGTTCAACAGTAA
Protein
MQYNGFHNQHPIFRPQPDRNEQFGKIPYIKGLPDFGGPKNFGPRPNMMNKNFRPRNDFNEVKNDYNTKNDGNQNDFGGPKQFRPRNNFNNGNQPPKKNNFNGDKSPGNMQYGNKNDFGGPKQQNYNKNYGPKTYNNQNCYGNEQPEFIPRQSYSPNSVQHSLNDRKLQSKKAKHPGDSLLKPMWDLTKLEPIQKDFYKPHVNVDARSEDDVQRFRVTKEITVSGNNIPRPNQLFEEGNFPDHIMSTIKEQPNLIEPTGIQAQGWPIALSGRDMVGIASTGSGKTLAYMLPAAVHIVHQQRIQRGDGPIALILAPTRELAQQIQSVAQAYSARGCIRNTCLFGGSPKGPQARDLERGVEIVIATPGRLIDFLERGTTNLRRCTYLVLDEADRMLDMGFEPQIRKIIEQIRPDRQVLMWSATWPKEIQALAEDFLTDYIKVNIGSLNLSANNNIKQIIEICEEHEKEVKLTNLLKEIASEKENKVIVFVETKKKVDDIARAVRRGGYKALAIHGDKSQPERDAVLTEFRNGSTTILIATDVAARGLDVEDVKFVVNFDYPNSSEDYIHRIGRTGRCQQSGTAYTYFTSGDCRQARGLLAVLRETGQNPPTKLADMARNNNNNSGRNRWQQRKDKNSGSNSPQQMSNQWNNKMANGSIEDGQNMYQNRSMNQQAPRFSNQNQGIQSYRQNNYQKQVPFPSPNVFESMGNYQGGYNNGYQNTYNQNGYGQRPFNNQRNNGQQYRNNGTNGNKASGSGSSFGSPPPHFATPPHYAQVHPHHQDMLGGKFYGSGVGGGGPCGYQAAVGAGVPYPHLPPGPLLYAAPVQQ
Summary
Similarity
Belongs to the DEAD box helicase family.
Uniprot
EMBL
BABH01011934
ODYU01011599
SOQ57440.1
GAIX01004468
JAA88092.1
KQ459590
+ More
KPI97449.1 JTDY01003416 KOB69589.1 AGBW02008593 OWR52969.1 GQ452007 ACV83780.1 KQ460205 KPJ16839.1 NNAY01004047 OXU18442.1 AAZX01007814 AK417733 BAN20948.1 GANO01000504 JAB59367.1 UFQS01000315 UFQT01000315 SSX02744.1 SSX23116.1
KPI97449.1 JTDY01003416 KOB69589.1 AGBW02008593 OWR52969.1 GQ452007 ACV83780.1 KQ460205 KPJ16839.1 NNAY01004047 OXU18442.1 AAZX01007814 AK417733 BAN20948.1 GANO01000504 JAB59367.1 UFQS01000315 UFQT01000315 SSX02744.1 SSX23116.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
PDB
4A4D
E-value=1.957e-97,
Score=911
Ontologies
GO
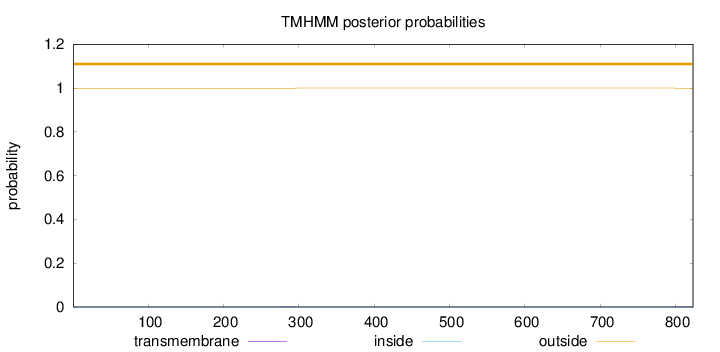
Topology
Length:
823
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01516
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00042
outside
1 - 823
Population Genetic Test Statistics
Pi
27.319142
Theta
146.108453
Tajima's D
-2.092154
CLR
4320.663781
CSRT
0.00974951252437378
Interpretation
Possibly Positive selection
