Gene
KWMTBOMO06704
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.867
Sequence
CDS
ATGACCGTCCAAGTACAGAAAGACCAGACGAGATCAATCCAGTTGCGCAGAGGTATAAGACAGGGGGATATTATATCCCCGAAACTATTTACAAGCGCACTGGAGGACGTCTTCAAGACTCTCGACTGGAACGGACGCGGCATTAACATAAACGGCGAGTACATTTCACATCTTCGCTTTGCCGATGACATCGTCATCATGGCGGTATCGCTGCAGGACCTACAAGAAAAGCTGTACAGCCTTAATACTGCATCTCAACGGGTAGATAAGACCAAGATAATATTTAACGGAAACGTCATTCCGAGACCAATCGATGTTGATGGTACACCTGAGGAAGTGTATGTCTACCTGGCAGAGACCCTACAACTAGGTAGAAATAATTTTGAGAAGGAAGTGATCAGAAGGATTCAGTTGGGCTGGGCAGCATTTGGGAAGCTTCGTCAGGTCTTTTCGTCACCCATACCACAATGTCTGAAGACAAATGTCTATAATCAGTGCGTCCTACCCGTGCTTACCTACGGAGCCGAAACGTGGACGCTGACCGTGCGACTAGTCTATCAACCAAAAATTACTCAGCGAGCTATAGAGCGAGCTATGCTTGGTTTATCATTGCGAGACCGAATACGTAATGAAGTTATTCGGGCAAAAACCAAGGTGATCGATATAGCTTGCAGAGTTAGTAAGCTGAAGTGGCAGTGGGCCGGTCACATATGTCGCAGAACCGATAACCTTTGGGGTAGACGTATTCTGGAGTGGAGACCGCGAACCGGCAAACGGAGTGTAGGACATCCTCCTGCTATTTGGAGCGACGATATGCGCAAGGCAGCTGGCAGTAACTGGTTGCATAGAACCGAAGATCGGGTTCAGTGGCGAGCTTTGGGGGAGGCCTATGTCCAGCAGTGGACTGCCACAGGCTGA
Protein
MTVQVQKDQTRSIQLRRGIRQGDIISPKLFTSALEDVFKTLDWNGRGININGEYISHLRFADDIVIMAVSLQDLQEKLYSLNTASQRVDKTKIIFNGNVIPRPIDVDGTPEEVYVYLAETLQLGRNNFEKEVIRRIQLGWAAFGKLRQVFSSPIPQCLKTNVYNQCVLPVLTYGAETWTLTVRLVYQPKITQRAIERAMLGLSLRDRIRNEVIRAKTKVIDIACRVSKLKWQWAGHICRRTDNLWGRRILEWRPRTGKRSVGHPPAIWSDDMRKAAGSNWLHRTEDRVQWRALGEAYVQQWTATG
Summary
Similarity
Belongs to the ATP-dependent DNA ligase family.
Uniprot
D7F157
D7F165
D7F166
A4KWF4
A4KWF2
A4KWG0
+ More
D7F164 A0A3S2TAR4 A0A3S2KX72 D7F160 D7F159 D7F158 A0A2W1BMB6 A0A3S2LUJ5 A0A2W1BLC9 W4YXJ1 A0A3S2TRE0 A0A2G8JFM9 A0A1E1X2U1 A0A2G8K4F2 A0A023EXV2 A0A2G8KES6 A0A2W1B5F8 A0A2W1BCS2 W4YXJ0 D7F177 D7F176 A0A2H1VY30 A0A016U3R5 D7F179 A0A2W1BSQ8 A0A2W1BDX2 D7F178 A0A016TIA8 A0A016WEQ1 A0A2W1BZV1 A0A016S795 A0A016WA79 A0A069DX96 A0A016S1D2 A0A023EWU8 A0A016S5B2 A0A016VVN3 A0A016W591 A0A224XSN7 A0A0K0FAW8 A0A0P4VWD0 A0A016V6V0 A0A016T7J7 A0A016UMK7 A0A016RXU5 A0A016RYK0 A0A016V7H3 A0A016VV56 A0A023GMF8 A0A016UTS3 A0A016WPY4 A0A016UWI5 A0A016SUK5 A0A016WCB8 A0A016ULU9 A0A131YK20 A0A016S403 A0A016U784 A0A016WKV3 A0A016UJL8 A0A016S949 A0A016TUG1 A0A016WMQ0 A0A016WC44 A0A2H1VS24 A0A069DZM5 A0A016WD35 A0A016USZ5 A0A016TQ81 A0A016VDZ6 A0A016WES8 A0A016WCG1 A0A016WD08 A0A016VBB9 A0A016VGM4 A0A016VN75 A0A016UUF2 A0A016VTY8 A0A016VMK8 A0A016SSP6 A0A016UR05 A0A016VW83 A0A016UGM6 A0A016TXK2
D7F164 A0A3S2TAR4 A0A3S2KX72 D7F160 D7F159 D7F158 A0A2W1BMB6 A0A3S2LUJ5 A0A2W1BLC9 W4YXJ1 A0A3S2TRE0 A0A2G8JFM9 A0A1E1X2U1 A0A2G8K4F2 A0A023EXV2 A0A2G8KES6 A0A2W1B5F8 A0A2W1BCS2 W4YXJ0 D7F177 D7F176 A0A2H1VY30 A0A016U3R5 D7F179 A0A2W1BSQ8 A0A2W1BDX2 D7F178 A0A016TIA8 A0A016WEQ1 A0A2W1BZV1 A0A016S795 A0A016WA79 A0A069DX96 A0A016S1D2 A0A023EWU8 A0A016S5B2 A0A016VVN3 A0A016W591 A0A224XSN7 A0A0K0FAW8 A0A0P4VWD0 A0A016V6V0 A0A016T7J7 A0A016UMK7 A0A016RXU5 A0A016RYK0 A0A016V7H3 A0A016VV56 A0A023GMF8 A0A016UTS3 A0A016WPY4 A0A016UWI5 A0A016SUK5 A0A016WCB8 A0A016ULU9 A0A131YK20 A0A016S403 A0A016U784 A0A016WKV3 A0A016UJL8 A0A016S949 A0A016TUG1 A0A016WMQ0 A0A016WC44 A0A2H1VS24 A0A069DZM5 A0A016WD35 A0A016USZ5 A0A016TQ81 A0A016VDZ6 A0A016WES8 A0A016WCG1 A0A016WD08 A0A016VBB9 A0A016VGM4 A0A016VN75 A0A016UUF2 A0A016VTY8 A0A016VMK8 A0A016SSP6 A0A016UR05 A0A016VW83 A0A016UGM6 A0A016TXK2
Pubmed
EMBL
FJ265542
ADI61810.1
FJ265550
ADI61818.1
FJ265551
ADI61819.1
+ More
EF113399 EF113400 ABO45233.1 EF113398 EF113401 ABO45231.1 EF113402 ABO45239.1 FJ265549 ADI61817.1 RSAL01002362 RVE40503.1 RSAL01002130 RVE40575.1 FJ265545 ADI61813.1 FJ265544 ADI61812.1 FJ265543 ADI61811.1 KZ150072 PZC74016.1 RSAL01000007 RVE54059.1 KZ149985 PZC75698.1 AAGJ04144474 RSAL01000016 RVE53061.1 MRZV01002136 PIK34564.1 GFAC01005640 JAT93548.1 MRZV01000899 PIK42833.1 GBBI01004876 JAC13836.1 MRZV01000640 PIK46507.1 KZ150317 PZC71439.1 KZ150485 PZC70710.1 AAGJ04050748 FJ265562 ADI61830.1 FJ265561 ADI61829.1 ODYU01004982 SOQ45402.1 JARK01001396 EYC09581.1 FJ265564 ADI61832.1 KZ149912 PZC78089.1 KZ150108 PZC73382.1 FJ265563 ADI61831.1 JARK01001434 EYC02684.1 JARK01000347 EYC38051.1 KZ149907 PZC78256.1 JARK01001621 EYB86109.1 JARK01000460 EYC36749.1 GBGD01000453 JAC88436.1 JARK01001655 EYB84321.1 GBBI01004877 JAC13835.1 JARK01001626 EYB85833.1 JARK01001340 EYC30828.1 JARK01000807 EYC34984.1 GFTR01005437 JAW10989.1 GDKW01001596 JAI54999.1 JARK01001351 EYC23394.1 JARK01001736 JARK01001462 JARK01001379 JARK01001338 EYB80775.1 EYB98943.1 EYC13646.1 EYC33347.1 JARK01001370 EYC16151.1 JARK01001677 EYB83168.1 EYB83167.1 EYC23395.1 EYC31295.1 GBBM01001358 JAC34060.1 JARK01001362 EYC18844.1 JARK01000173 EYC41342.1 EYC18843.1 JARK01001512 EYB94036.1 JARK01000388 EYC37469.1 EYC16150.1 GEDV01010296 JAP78261.1 JARK01001639 EYB85201.1 JARK01001389 EYC11015.1 JARK01000224 EYC40226.1 JARK01001372 EYC15539.1 JARK01001608 EYB86877.1 JARK01001414 JARK01001412 EYC06128.1 EYC06440.1 JARK01000189 EYC40935.1 JARK01000449 EYC36872.1 ODYU01004088 SOQ43598.1 GBGD01000380 JAC88509.1 JARK01000384 EYC37506.1 JARK01001364 EYC18031.1 JARK01001423 EYC04578.1 JARK01001348 EYC25257.1 EYC37503.1 EYC37504.1 EYC37505.1 JARK01001349 EYC24735.1 JARK01001346 EYC26784.1 JARK01001344 EYC28218.1 JARK01001361 EYC19034.1 JARK01001341 EYC30223.1 JARK01001343 EYC28635.1 JARK01001517 EYB93530.1 JARK01001367 EYC17351.1 EYC31302.1 JARK01001378 EYC13977.1 JARK01001406 EYC07496.1
EF113399 EF113400 ABO45233.1 EF113398 EF113401 ABO45231.1 EF113402 ABO45239.1 FJ265549 ADI61817.1 RSAL01002362 RVE40503.1 RSAL01002130 RVE40575.1 FJ265545 ADI61813.1 FJ265544 ADI61812.1 FJ265543 ADI61811.1 KZ150072 PZC74016.1 RSAL01000007 RVE54059.1 KZ149985 PZC75698.1 AAGJ04144474 RSAL01000016 RVE53061.1 MRZV01002136 PIK34564.1 GFAC01005640 JAT93548.1 MRZV01000899 PIK42833.1 GBBI01004876 JAC13836.1 MRZV01000640 PIK46507.1 KZ150317 PZC71439.1 KZ150485 PZC70710.1 AAGJ04050748 FJ265562 ADI61830.1 FJ265561 ADI61829.1 ODYU01004982 SOQ45402.1 JARK01001396 EYC09581.1 FJ265564 ADI61832.1 KZ149912 PZC78089.1 KZ150108 PZC73382.1 FJ265563 ADI61831.1 JARK01001434 EYC02684.1 JARK01000347 EYC38051.1 KZ149907 PZC78256.1 JARK01001621 EYB86109.1 JARK01000460 EYC36749.1 GBGD01000453 JAC88436.1 JARK01001655 EYB84321.1 GBBI01004877 JAC13835.1 JARK01001626 EYB85833.1 JARK01001340 EYC30828.1 JARK01000807 EYC34984.1 GFTR01005437 JAW10989.1 GDKW01001596 JAI54999.1 JARK01001351 EYC23394.1 JARK01001736 JARK01001462 JARK01001379 JARK01001338 EYB80775.1 EYB98943.1 EYC13646.1 EYC33347.1 JARK01001370 EYC16151.1 JARK01001677 EYB83168.1 EYB83167.1 EYC23395.1 EYC31295.1 GBBM01001358 JAC34060.1 JARK01001362 EYC18844.1 JARK01000173 EYC41342.1 EYC18843.1 JARK01001512 EYB94036.1 JARK01000388 EYC37469.1 EYC16150.1 GEDV01010296 JAP78261.1 JARK01001639 EYB85201.1 JARK01001389 EYC11015.1 JARK01000224 EYC40226.1 JARK01001372 EYC15539.1 JARK01001608 EYB86877.1 JARK01001414 JARK01001412 EYC06128.1 EYC06440.1 JARK01000189 EYC40935.1 JARK01000449 EYC36872.1 ODYU01004088 SOQ43598.1 GBGD01000380 JAC88509.1 JARK01000384 EYC37506.1 JARK01001364 EYC18031.1 JARK01001423 EYC04578.1 JARK01001348 EYC25257.1 EYC37503.1 EYC37504.1 EYC37505.1 JARK01001349 EYC24735.1 JARK01001346 EYC26784.1 JARK01001344 EYC28218.1 JARK01001361 EYC19034.1 JARK01001341 EYC30223.1 JARK01001343 EYC28635.1 JARK01001517 EYB93530.1 JARK01001367 EYC17351.1 EYC31302.1 JARK01001378 EYC13977.1 JARK01001406 EYC07496.1
Proteomes
Pfam
Interpro
IPR005135
Endo/exonuclease/phosphatase
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR036084 Ser_inhib-like_sf
IPR002919 TIL_dom
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR012340 NA-bd_OB-fold
IPR016059 DNA_ligase_ATP-dep_CS
IPR012310 DNA_ligase_ATP-dep_cent
IPR021536 DNA_ligase_IV_dom
IPR029710 LIG4
IPR000977 DNA_ligase_ATP-dep
IPR012308 DNA_ligase_ATP-dep_N
IPR036599 DNA_ligase_N_sf
IPR012309 DNA_ligase_ATP-dep_C
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR036084 Ser_inhib-like_sf
IPR002919 TIL_dom
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR012340 NA-bd_OB-fold
IPR016059 DNA_ligase_ATP-dep_CS
IPR012310 DNA_ligase_ATP-dep_cent
IPR021536 DNA_ligase_IV_dom
IPR029710 LIG4
IPR000977 DNA_ligase_ATP-dep
IPR012308 DNA_ligase_ATP-dep_N
IPR036599 DNA_ligase_N_sf
IPR012309 DNA_ligase_ATP-dep_C
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
ProteinModelPortal
D7F157
D7F165
D7F166
A4KWF4
A4KWF2
A4KWG0
+ More
D7F164 A0A3S2TAR4 A0A3S2KX72 D7F160 D7F159 D7F158 A0A2W1BMB6 A0A3S2LUJ5 A0A2W1BLC9 W4YXJ1 A0A3S2TRE0 A0A2G8JFM9 A0A1E1X2U1 A0A2G8K4F2 A0A023EXV2 A0A2G8KES6 A0A2W1B5F8 A0A2W1BCS2 W4YXJ0 D7F177 D7F176 A0A2H1VY30 A0A016U3R5 D7F179 A0A2W1BSQ8 A0A2W1BDX2 D7F178 A0A016TIA8 A0A016WEQ1 A0A2W1BZV1 A0A016S795 A0A016WA79 A0A069DX96 A0A016S1D2 A0A023EWU8 A0A016S5B2 A0A016VVN3 A0A016W591 A0A224XSN7 A0A0K0FAW8 A0A0P4VWD0 A0A016V6V0 A0A016T7J7 A0A016UMK7 A0A016RXU5 A0A016RYK0 A0A016V7H3 A0A016VV56 A0A023GMF8 A0A016UTS3 A0A016WPY4 A0A016UWI5 A0A016SUK5 A0A016WCB8 A0A016ULU9 A0A131YK20 A0A016S403 A0A016U784 A0A016WKV3 A0A016UJL8 A0A016S949 A0A016TUG1 A0A016WMQ0 A0A016WC44 A0A2H1VS24 A0A069DZM5 A0A016WD35 A0A016USZ5 A0A016TQ81 A0A016VDZ6 A0A016WES8 A0A016WCG1 A0A016WD08 A0A016VBB9 A0A016VGM4 A0A016VN75 A0A016UUF2 A0A016VTY8 A0A016VMK8 A0A016SSP6 A0A016UR05 A0A016VW83 A0A016UGM6 A0A016TXK2
D7F164 A0A3S2TAR4 A0A3S2KX72 D7F160 D7F159 D7F158 A0A2W1BMB6 A0A3S2LUJ5 A0A2W1BLC9 W4YXJ1 A0A3S2TRE0 A0A2G8JFM9 A0A1E1X2U1 A0A2G8K4F2 A0A023EXV2 A0A2G8KES6 A0A2W1B5F8 A0A2W1BCS2 W4YXJ0 D7F177 D7F176 A0A2H1VY30 A0A016U3R5 D7F179 A0A2W1BSQ8 A0A2W1BDX2 D7F178 A0A016TIA8 A0A016WEQ1 A0A2W1BZV1 A0A016S795 A0A016WA79 A0A069DX96 A0A016S1D2 A0A023EWU8 A0A016S5B2 A0A016VVN3 A0A016W591 A0A224XSN7 A0A0K0FAW8 A0A0P4VWD0 A0A016V6V0 A0A016T7J7 A0A016UMK7 A0A016RXU5 A0A016RYK0 A0A016V7H3 A0A016VV56 A0A023GMF8 A0A016UTS3 A0A016WPY4 A0A016UWI5 A0A016SUK5 A0A016WCB8 A0A016ULU9 A0A131YK20 A0A016S403 A0A016U784 A0A016WKV3 A0A016UJL8 A0A016S949 A0A016TUG1 A0A016WMQ0 A0A016WC44 A0A2H1VS24 A0A069DZM5 A0A016WD35 A0A016USZ5 A0A016TQ81 A0A016VDZ6 A0A016WES8 A0A016WCG1 A0A016WD08 A0A016VBB9 A0A016VGM4 A0A016VN75 A0A016UUF2 A0A016VTY8 A0A016VMK8 A0A016SSP6 A0A016UR05 A0A016VW83 A0A016UGM6 A0A016TXK2
Ontologies
PANTHER
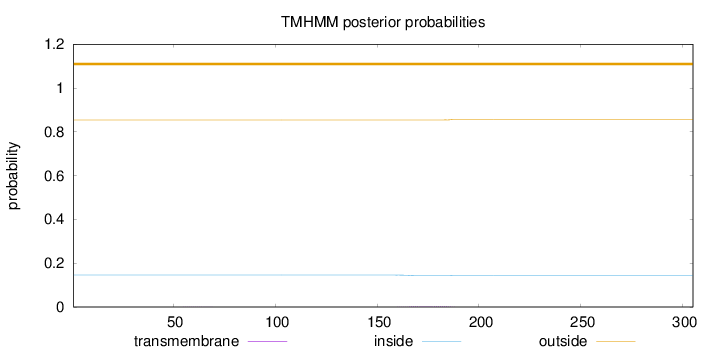
Topology
Length:
305
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06876
Exp number, first 60 AAs:
0.00126
Total prob of N-in:
0.14609
outside
1 - 305
Population Genetic Test Statistics
Pi
247.368203
Theta
199.270558
Tajima's D
0.759736
CLR
28.736285
CSRT
0.585220738963052
Interpretation
Uncertain
