Pre Gene Modal
BGIBMGA011756
Annotation
uncharacterized_LOC101746868_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.424 Mitochondrial Reliability : 1.847
Sequence
CDS
ATGGCTACCGGTACTGTAATAAAATATGTTGGAAGAACGACTGACTTTAAAGGAAAATCACTTTGGGAAATCGTTGGTAGCCTAAAGAATTTTGGTGTAGGCAGGATTATTGTCAGATCTGTTTTCTTGCGTTACCCAGAACCTAGTTTTATGAAAATCGTGAAAGTGCAAACTTGCCCCGACGAGGAAAGGAGAAGAGTACGTGTTTGGGTTGAGAAAACATTCAGAGGACAAAAACTACCCAATATTACAGAAATATATCGCACATCATACAAAACTGATTATCAGCTTATCCCCAAAAGAGAAGAAACTAGGTTGTTAGAGTCTATAAACACCATTGGAAATAAGGCAGATGTCATATTACCGAATTCTATTGAAATGCCACCATTGATGAAACTGTTTATCATTAAAGATCATGAAAAGAAAGGACTAGAAACCTCCAAAGACTTCATGATGCCCCTCAGTTATAATCAAAGTCCAAATAGAACAAACAAGATAGCAAAAGCCAATGAACAACCCACAATAAAGTTTACTATGGGATTAGGAAAACCAGTCAGTCCCTCCCTGTATGATGGTATCCCTCTATAA
Protein
MATGTVIKYVGRTTDFKGKSLWEIVGSLKNFGVGRIIVRSVFLRYPEPSFMKIVKVQTCPDEERRRVRVWVEKTFRGQKLPNITEIYRTSYKTDYQLIPKREETRLLESINTIGNKADVILPNSIEMPPLMKLFIIKDHEKKGLETSKDFMMPLSYNQSPNRTNKIAKANEQPTIKFTMGLGKPVSPSLYDGIPL
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JQF0
Q2F5Y4
A0A194PWQ1
A0A194RHN2
A0A2H1X0K5
A0A2A4JXG5
+ More
A0A1E1WBJ1 A0A212FGY5 A0A182WWJ2 A0A182UWL2 A0A182L3K7 A0A182I6S3 A0A182UCE2 A0A182JKX7 A0A182Y2C9 A0A182PII0 A0A182LY79 Q7QIM6 Q17JQ1 A0A182WBJ3 A0A084WEZ6 A0A182JP78 A0A067RKK9 A0A182N995 A0A1S4F083 Q17JQ3 B0WRY2 A0A182RJM2 A0A182QUR8 A0A1Q3FBS4 A0A0K8TRT1 A0A2M4BZI8 A0A1W4XAH3 A0A2P8Z9R0 A0A182FGU0 A0A0K8WJI4 A0A034WC75 A0A0L0CCU0 A0A1A9UJ34 A0A2M4C145 T1DKM6 W8C724 A0A2M3ZC65 A0A0A1WYM1 A0A2M4ASC1 A0A2M4C134 Q29EJ9 B4H243 A0A1B0GAS4 T1P8T4 A0A1B6D1J7 A0A2M3ZCN2 A0A1B0A6F1 A0A1I8PC70 B4J0T8 B4L9L5 A0A3B0JYB7 A0A1L8EH17 W5JNY6 U5ENN2 Q9VV39 A0A0M4EPE3 A0A1A9X0L2 B3M5J9 B4LI28 A0A1B0CJJ4 B4QMD6 A0A1A9YKP0 A0A1B0BY31 A0A1W4UHL4 A0A310SSW1 B3NIB7 K7J867 B4HJL3 B4PHI7 J3JTS2 B4MX85 A0A1Y1L490 A0A1J1I8H9 A0A0C9QZQ5 A0A139WM14 A0A1L8E1I0 A0A232EGJ9 A0A2A3EN28 A0A0M9A836 A0A023F7Y5 A0A1S3D1D4 A0A087ZSC5 A0A224XQM8 A0A0V0G915 A0A0P4VTH5 R4FKS5 A0A1L8E149 A0A0J7L2J2 A0A1B6EQ94 A0A026X0U0 A0A3L8DSM2 A0A158NU59
A0A1E1WBJ1 A0A212FGY5 A0A182WWJ2 A0A182UWL2 A0A182L3K7 A0A182I6S3 A0A182UCE2 A0A182JKX7 A0A182Y2C9 A0A182PII0 A0A182LY79 Q7QIM6 Q17JQ1 A0A182WBJ3 A0A084WEZ6 A0A182JP78 A0A067RKK9 A0A182N995 A0A1S4F083 Q17JQ3 B0WRY2 A0A182RJM2 A0A182QUR8 A0A1Q3FBS4 A0A0K8TRT1 A0A2M4BZI8 A0A1W4XAH3 A0A2P8Z9R0 A0A182FGU0 A0A0K8WJI4 A0A034WC75 A0A0L0CCU0 A0A1A9UJ34 A0A2M4C145 T1DKM6 W8C724 A0A2M3ZC65 A0A0A1WYM1 A0A2M4ASC1 A0A2M4C134 Q29EJ9 B4H243 A0A1B0GAS4 T1P8T4 A0A1B6D1J7 A0A2M3ZCN2 A0A1B0A6F1 A0A1I8PC70 B4J0T8 B4L9L5 A0A3B0JYB7 A0A1L8EH17 W5JNY6 U5ENN2 Q9VV39 A0A0M4EPE3 A0A1A9X0L2 B3M5J9 B4LI28 A0A1B0CJJ4 B4QMD6 A0A1A9YKP0 A0A1B0BY31 A0A1W4UHL4 A0A310SSW1 B3NIB7 K7J867 B4HJL3 B4PHI7 J3JTS2 B4MX85 A0A1Y1L490 A0A1J1I8H9 A0A0C9QZQ5 A0A139WM14 A0A1L8E1I0 A0A232EGJ9 A0A2A3EN28 A0A0M9A836 A0A023F7Y5 A0A1S3D1D4 A0A087ZSC5 A0A224XQM8 A0A0V0G915 A0A0P4VTH5 R4FKS5 A0A1L8E149 A0A0J7L2J2 A0A1B6EQ94 A0A026X0U0 A0A3L8DSM2 A0A158NU59
Pubmed
19121390
26354079
22118469
20966253
25244985
12364791
+ More
14747013 17210077 17510324 24438588 24845553 26369729 29403074 25348373 26108605 24495485 25830018 15632085 17994087 25315136 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20075255 17550304 22516182 23537049 28004739 18362917 19820115 28648823 25474469 27129103 24508170 30249741 21347285
14747013 17210077 17510324 24438588 24845553 26369729 29403074 25348373 26108605 24495485 25830018 15632085 17994087 25315136 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20075255 17550304 22516182 23537049 28004739 18362917 19820115 28648823 25474469 27129103 24508170 30249741 21347285
EMBL
BABH01011943
DQ311288
ABD36233.2
KQ459590
KPI97443.1
KQ460205
+ More
KPJ16835.1 ODYU01012477 SOQ58807.1 NWSH01000425 PCG76489.1 GDQN01006718 JAT84336.1 AGBW02008593 OWR52973.1 APCN01003693 AXCM01000934 AAAB01008807 EAA04429.4 CH477231 EAT46923.1 ATLV01023266 KE525341 KFB48790.1 KK852621 KDR20025.1 EAT46921.1 DS232062 EDS33592.1 AXCN02000558 GFDL01010014 JAV25031.1 GDAI01000529 JAI17074.1 GGFJ01009346 MBW58487.1 PYGN01000135 PSN53237.1 GDHF01000998 JAI51316.1 GAKP01007247 JAC51705.1 JRES01000577 KNC30065.1 GGFJ01009884 MBW59025.1 GAMD01000876 JAB00715.1 GAMC01003884 JAC02672.1 GGFM01005405 MBW26156.1 GBXI01010672 JAD03620.1 GGFK01010385 MBW43706.1 GGFJ01009885 MBW59026.1 CH379070 EAL30061.1 CH479203 EDW30395.1 CCAG010015502 KA644540 AFP59169.1 GEDC01028049 GEDC01017719 JAS09249.1 JAS19579.1 GGFM01005553 MBW26304.1 CH916366 EDV95759.1 CH933816 EDW17390.1 OUUW01000012 SPP87067.1 GFDG01000854 JAV17945.1 ADMH02000528 ETN66097.1 GANO01004004 JAB55867.1 AE014296 BT099501 AAF49483.2 ACU24741.1 CP012525 ALC43967.1 CH902618 EDV39609.1 CH940647 EDW68572.1 AJWK01014674 CM000363 CM002912 EDX10705.1 KMZ00022.1 JXJN01022474 KQ760382 OAD60738.1 CH954178 EDV52273.1 CH480815 EDW41740.1 CM000159 EDW95425.1 BT126631 KB631760 AEE61594.1 ERL85892.1 CH963876 EDW76918.1 GEZM01068780 JAV66775.1 CVRI01000043 CRK95876.1 GBYB01009254 JAG79021.1 KQ971319 KYB28924.1 GFDF01001662 JAV12422.1 NNAY01004755 OXU17461.1 KZ288204 PBC33165.1 KQ435714 KOX79170.1 GBBI01001345 JAC17367.1 GFTR01003032 JAW13394.1 GECL01002343 JAP03781.1 GDKW01001305 JAI55290.1 ACPB03012525 GAHY01001718 JAA75792.1 GFDF01001675 JAV12409.1 LBMM01001124 KMQ96878.1 GECZ01029658 JAS40111.1 KK107046 EZA61688.1 QOIP01000004 RLU23420.1 ADTU01026245
KPJ16835.1 ODYU01012477 SOQ58807.1 NWSH01000425 PCG76489.1 GDQN01006718 JAT84336.1 AGBW02008593 OWR52973.1 APCN01003693 AXCM01000934 AAAB01008807 EAA04429.4 CH477231 EAT46923.1 ATLV01023266 KE525341 KFB48790.1 KK852621 KDR20025.1 EAT46921.1 DS232062 EDS33592.1 AXCN02000558 GFDL01010014 JAV25031.1 GDAI01000529 JAI17074.1 GGFJ01009346 MBW58487.1 PYGN01000135 PSN53237.1 GDHF01000998 JAI51316.1 GAKP01007247 JAC51705.1 JRES01000577 KNC30065.1 GGFJ01009884 MBW59025.1 GAMD01000876 JAB00715.1 GAMC01003884 JAC02672.1 GGFM01005405 MBW26156.1 GBXI01010672 JAD03620.1 GGFK01010385 MBW43706.1 GGFJ01009885 MBW59026.1 CH379070 EAL30061.1 CH479203 EDW30395.1 CCAG010015502 KA644540 AFP59169.1 GEDC01028049 GEDC01017719 JAS09249.1 JAS19579.1 GGFM01005553 MBW26304.1 CH916366 EDV95759.1 CH933816 EDW17390.1 OUUW01000012 SPP87067.1 GFDG01000854 JAV17945.1 ADMH02000528 ETN66097.1 GANO01004004 JAB55867.1 AE014296 BT099501 AAF49483.2 ACU24741.1 CP012525 ALC43967.1 CH902618 EDV39609.1 CH940647 EDW68572.1 AJWK01014674 CM000363 CM002912 EDX10705.1 KMZ00022.1 JXJN01022474 KQ760382 OAD60738.1 CH954178 EDV52273.1 CH480815 EDW41740.1 CM000159 EDW95425.1 BT126631 KB631760 AEE61594.1 ERL85892.1 CH963876 EDW76918.1 GEZM01068780 JAV66775.1 CVRI01000043 CRK95876.1 GBYB01009254 JAG79021.1 KQ971319 KYB28924.1 GFDF01001662 JAV12422.1 NNAY01004755 OXU17461.1 KZ288204 PBC33165.1 KQ435714 KOX79170.1 GBBI01001345 JAC17367.1 GFTR01003032 JAW13394.1 GECL01002343 JAP03781.1 GDKW01001305 JAI55290.1 ACPB03012525 GAHY01001718 JAA75792.1 GFDF01001675 JAV12409.1 LBMM01001124 KMQ96878.1 GECZ01029658 JAS40111.1 KK107046 EZA61688.1 QOIP01000004 RLU23420.1 ADTU01026245
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000076407
+ More
UP000075903 UP000075882 UP000075840 UP000075902 UP000075880 UP000076408 UP000075885 UP000075883 UP000007062 UP000008820 UP000075920 UP000030765 UP000075881 UP000027135 UP000075884 UP000002320 UP000075900 UP000075886 UP000192223 UP000245037 UP000069272 UP000037069 UP000078200 UP000001819 UP000008744 UP000092444 UP000095301 UP000092445 UP000095300 UP000001070 UP000009192 UP000268350 UP000000673 UP000000803 UP000092553 UP000091820 UP000007801 UP000008792 UP000092461 UP000000304 UP000092443 UP000092460 UP000192221 UP000008711 UP000002358 UP000001292 UP000002282 UP000030742 UP000007798 UP000183832 UP000007266 UP000215335 UP000242457 UP000053105 UP000079169 UP000005203 UP000015103 UP000036403 UP000053097 UP000279307 UP000005205
UP000075903 UP000075882 UP000075840 UP000075902 UP000075880 UP000076408 UP000075885 UP000075883 UP000007062 UP000008820 UP000075920 UP000030765 UP000075881 UP000027135 UP000075884 UP000002320 UP000075900 UP000075886 UP000192223 UP000245037 UP000069272 UP000037069 UP000078200 UP000001819 UP000008744 UP000092444 UP000095301 UP000092445 UP000095300 UP000001070 UP000009192 UP000268350 UP000000673 UP000000803 UP000092553 UP000091820 UP000007801 UP000008792 UP000092461 UP000000304 UP000092443 UP000092460 UP000192221 UP000008711 UP000002358 UP000001292 UP000002282 UP000030742 UP000007798 UP000183832 UP000007266 UP000215335 UP000242457 UP000053105 UP000079169 UP000005203 UP000015103 UP000036403 UP000053097 UP000279307 UP000005205
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9JQF0
Q2F5Y4
A0A194PWQ1
A0A194RHN2
A0A2H1X0K5
A0A2A4JXG5
+ More
A0A1E1WBJ1 A0A212FGY5 A0A182WWJ2 A0A182UWL2 A0A182L3K7 A0A182I6S3 A0A182UCE2 A0A182JKX7 A0A182Y2C9 A0A182PII0 A0A182LY79 Q7QIM6 Q17JQ1 A0A182WBJ3 A0A084WEZ6 A0A182JP78 A0A067RKK9 A0A182N995 A0A1S4F083 Q17JQ3 B0WRY2 A0A182RJM2 A0A182QUR8 A0A1Q3FBS4 A0A0K8TRT1 A0A2M4BZI8 A0A1W4XAH3 A0A2P8Z9R0 A0A182FGU0 A0A0K8WJI4 A0A034WC75 A0A0L0CCU0 A0A1A9UJ34 A0A2M4C145 T1DKM6 W8C724 A0A2M3ZC65 A0A0A1WYM1 A0A2M4ASC1 A0A2M4C134 Q29EJ9 B4H243 A0A1B0GAS4 T1P8T4 A0A1B6D1J7 A0A2M3ZCN2 A0A1B0A6F1 A0A1I8PC70 B4J0T8 B4L9L5 A0A3B0JYB7 A0A1L8EH17 W5JNY6 U5ENN2 Q9VV39 A0A0M4EPE3 A0A1A9X0L2 B3M5J9 B4LI28 A0A1B0CJJ4 B4QMD6 A0A1A9YKP0 A0A1B0BY31 A0A1W4UHL4 A0A310SSW1 B3NIB7 K7J867 B4HJL3 B4PHI7 J3JTS2 B4MX85 A0A1Y1L490 A0A1J1I8H9 A0A0C9QZQ5 A0A139WM14 A0A1L8E1I0 A0A232EGJ9 A0A2A3EN28 A0A0M9A836 A0A023F7Y5 A0A1S3D1D4 A0A087ZSC5 A0A224XQM8 A0A0V0G915 A0A0P4VTH5 R4FKS5 A0A1L8E149 A0A0J7L2J2 A0A1B6EQ94 A0A026X0U0 A0A3L8DSM2 A0A158NU59
A0A1E1WBJ1 A0A212FGY5 A0A182WWJ2 A0A182UWL2 A0A182L3K7 A0A182I6S3 A0A182UCE2 A0A182JKX7 A0A182Y2C9 A0A182PII0 A0A182LY79 Q7QIM6 Q17JQ1 A0A182WBJ3 A0A084WEZ6 A0A182JP78 A0A067RKK9 A0A182N995 A0A1S4F083 Q17JQ3 B0WRY2 A0A182RJM2 A0A182QUR8 A0A1Q3FBS4 A0A0K8TRT1 A0A2M4BZI8 A0A1W4XAH3 A0A2P8Z9R0 A0A182FGU0 A0A0K8WJI4 A0A034WC75 A0A0L0CCU0 A0A1A9UJ34 A0A2M4C145 T1DKM6 W8C724 A0A2M3ZC65 A0A0A1WYM1 A0A2M4ASC1 A0A2M4C134 Q29EJ9 B4H243 A0A1B0GAS4 T1P8T4 A0A1B6D1J7 A0A2M3ZCN2 A0A1B0A6F1 A0A1I8PC70 B4J0T8 B4L9L5 A0A3B0JYB7 A0A1L8EH17 W5JNY6 U5ENN2 Q9VV39 A0A0M4EPE3 A0A1A9X0L2 B3M5J9 B4LI28 A0A1B0CJJ4 B4QMD6 A0A1A9YKP0 A0A1B0BY31 A0A1W4UHL4 A0A310SSW1 B3NIB7 K7J867 B4HJL3 B4PHI7 J3JTS2 B4MX85 A0A1Y1L490 A0A1J1I8H9 A0A0C9QZQ5 A0A139WM14 A0A1L8E1I0 A0A232EGJ9 A0A2A3EN28 A0A0M9A836 A0A023F7Y5 A0A1S3D1D4 A0A087ZSC5 A0A224XQM8 A0A0V0G915 A0A0P4VTH5 R4FKS5 A0A1L8E149 A0A0J7L2J2 A0A1B6EQ94 A0A026X0U0 A0A3L8DSM2 A0A158NU59
PDB
6NF8
E-value=2.0379e-05,
Score=111
Ontologies
GO
PANTHER
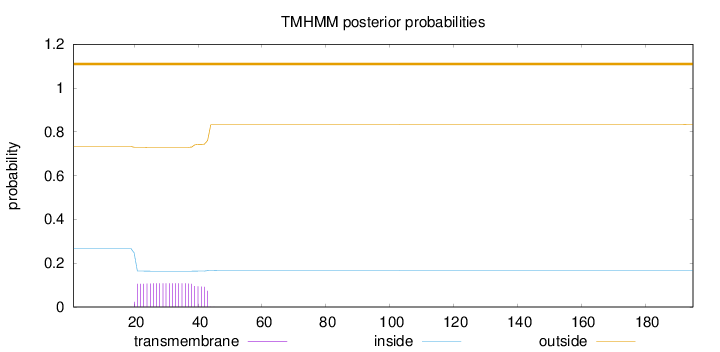
Topology
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.40221
Exp number, first 60 AAs:
2.40105
Total prob of N-in:
0.26725
outside
1 - 195
Population Genetic Test Statistics
Pi
18.422829
Theta
19.488606
Tajima's D
-0.210107
CLR
0.647054
CSRT
0.31668416579171
Interpretation
Uncertain
