Pre Gene Modal
BGIBMGA011770
Annotation
G_protein_pathway_suppressor_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.253
Sequence
CDS
ATGAACACAGCTGAACCAATGCAGGTTGATATTCCCCCAGAAGATAATGAAAACAATGAGACGGAATGCTACGTCGTTGAAAACCCCACCTTGGATTTGGAAACATACGCTGCGTCATATACTGGTTTTGCAAAACTTTATAGGCTCATGTTTGTAGCAGACCACTGTCCGTCATTGCGATTGGAAGCTTTGAAAATGGCCATATCCTATGTCATGACCACATATAACGTAAATCTGTATCACACACTGCATAAAAAACTCTCAGAAGCAGTGGCATCAGCAGGACTGCCTGATATAGCAGGTTCTCAAGATATTCCAGTCTTAGATACTATTTGGGTAGAATCAAAAACAAAAAAAGCTGCCATAAAACTGGAAAAGCTAGATACTGATTTGAAAAATTATAAAACAAATTCAATCAAAGAAAGTATCAGAAGAGGCCATGATGATTTGGGTGACCATTATCTAGATTGTGGAGATCTCACCAGTGCCTTGAAATGCTACTCGAGAGCTAGAGATTACTGCACAAGTGGAAAACATCTTGTCATGATGTGCTTGAATGTAGTTAAAGTTTCAGTATATCTTCAAAATTGGGCTCATGTTCTAAATTATGTATCTAAAGCTGAAGCGACCCCAGACTTTAATGAAATTCCCGGAAAAGACAGCAACCAGTCAATTTTAACTCGTTTAAAATGTGCAGCAGGATTAGCCGAGCTGGCAACGAAAAAGTATAAATCTGCTGCAAAACATTTTTTGGCTGCCAGCATTGATCATTGTGAATACCCAGAATTAATGAGCAGTAATAATGTTGCCATCTATGGTGGACTTTGCGCACTTGCTACATTTGATCGTTCAGAGTTGCAGAAACAAGTCATAGTTAGCAGTTCTTTTAAATTATTCTTAGAGCTAGAGCCCCAATTACGTGACATAATTTTCAAATTCTATGAATCCAAATATGCCTCGTGCTTACGGCTGTTGGATGAAATAAGGGATAATCTTTTGTTGGACATGTATTTAGCACCACATATAAATTCTTTATACATGCAAATTCGCAACAGGGCTTTGATTCAATACTTTAGCCCTTACTTGTCAGCTGATATGAAATTAATGGCGGCTGCATTTAACCGATCGGTGATTGCCTTAGAAGATGAACTAATGCAACTTATACTTGATGGGCAAATTCAGGCTAGGATTGATTCACATAATAAAATATTATATGCAAAAGATGTAGACCAGAGGAGCACAACATTTGAAAGAAGTTTAGCCATGGGAAAAGAATACCAACGGCGTACGTCAATGTTAATTTTGCGTGCTGCAATGCTCAAAAACCAAATCCATGTAAAGAATATTGGCAGAGACATAGGACCTCAAAGTGGAGAAGCTCCAGGCTCCAGTACTAGTTTGAACACTCGCACTTTCGATCCAGTACCTCTGTGA
Protein
MNTAEPMQVDIPPEDNENNETECYVVENPTLDLETYAASYTGFAKLYRLMFVADHCPSLRLEALKMAISYVMTTYNVNLYHTLHKKLSEAVASAGLPDIAGSQDIPVLDTIWVESKTKKAAIKLEKLDTDLKNYKTNSIKESIRRGHDDLGDHYLDCGDLTSALKCYSRARDYCTSGKHLVMMCLNVVKVSVYLQNWAHVLNYVSKAEATPDFNEIPGKDSNQSILTRLKCAAGLAELATKKYKSAAKHFLAASIDHCEYPELMSSNNVAIYGGLCALATFDRSELQKQVIVSSSFKLFLELEPQLRDIIFKFYESKYASCLRLLDEIRDNLLLDMYLAPHINSLYMQIRNRALIQYFSPYLSADMKLMAAAFNRSVIALEDELMQLILDGQIQARIDSHNKILYAKDVDQRSTTFERSLAMGKEYQRRTSMLILRAAMLKNQIHVKNIGRDIGPQSGEAPGSSTSLNTRTFDPVPL
Summary
Uniprot
Q2F5R2
A0A2A4K6X6
A0A2H1W709
S4Q0A8
A0A3S2LQQ0
A0A212FJ83
+ More
A0A194RHK1 A0A0L7LHE4 D6WF13 A0A2J7QSS0 A0A0T6B394 A0A2J7QSR7 A0A067RBT3 E2BWT6 A0A1Y1LAQ0 A0A1Y1LAP6 A0A1L8DW78 E2A7N4 E9I9P5 A0A195CKR1 A0A195EV85 A0A0J7KR61 A0A151I0R2 A0A2A3ESE3 A0A088AP80 A0A151J0G6 F4WMJ1 A0A1B6LTV4 A0A158NAE9 A0A151X4Y1 A0A1B0DBQ5 E0VVX5 A0A0L7QN62 A0A026WG97 A0A1W4WN96 A0A154PNT6 T1J1H8 A0A2R7W465 A0A310STA3 R4WTT0 A0A1B6EQ85 A0A1B6E3J4 A0A1B0CY66 A0A0V0G9E2 A0A069DTZ7 A0A224XFP5 A0A0A9X3K0 A0A182JVZ8 A0A182PTV4 A0A0K8SNE6 A0A182UWY4 A0A182KNP6 Q7Q7Y5 A0A182HQL1 A0A232FC43 A0A182J1J0 A0A087URG7 A0A0L0C7W4 A0A0P4VQI5 A0A1I8NY35 A0A182SKX2 R4G461 A0A023F9C2 A0A182YR54 A0A182XEG8 A0A182MQR2 A0A182WM19 A0A1I8N4T2 A0A182RE50 A0A182TD49 A0A182Q6R4 A0A182N1Y9 T1PC05 A0A1B0C6Q0 A0A1A9ZDX0 A0A146MFQ9 A0A1A9WLL5 A0A084WE14 A0A0P6IY33 A0A034WPS5 A0A0K8TVX2 A0A0A1XQY2 A0A1A9XJV5 A0A1A9UKQ9 A0A182H2Z8 W8BBQ2 A0A2R5LEC8 A0A1B0GF73 W5JV58 A0A2M4AKP4 K1RS36 A0A2M4AJT4 A0A1Q3G2N0 A0A210QT24 A0A1S3JDM1 A0A224YK68 A0A131YJI1 A0A131YE58 A0A1J1I5K2 L7MGY1
A0A194RHK1 A0A0L7LHE4 D6WF13 A0A2J7QSS0 A0A0T6B394 A0A2J7QSR7 A0A067RBT3 E2BWT6 A0A1Y1LAQ0 A0A1Y1LAP6 A0A1L8DW78 E2A7N4 E9I9P5 A0A195CKR1 A0A195EV85 A0A0J7KR61 A0A151I0R2 A0A2A3ESE3 A0A088AP80 A0A151J0G6 F4WMJ1 A0A1B6LTV4 A0A158NAE9 A0A151X4Y1 A0A1B0DBQ5 E0VVX5 A0A0L7QN62 A0A026WG97 A0A1W4WN96 A0A154PNT6 T1J1H8 A0A2R7W465 A0A310STA3 R4WTT0 A0A1B6EQ85 A0A1B6E3J4 A0A1B0CY66 A0A0V0G9E2 A0A069DTZ7 A0A224XFP5 A0A0A9X3K0 A0A182JVZ8 A0A182PTV4 A0A0K8SNE6 A0A182UWY4 A0A182KNP6 Q7Q7Y5 A0A182HQL1 A0A232FC43 A0A182J1J0 A0A087URG7 A0A0L0C7W4 A0A0P4VQI5 A0A1I8NY35 A0A182SKX2 R4G461 A0A023F9C2 A0A182YR54 A0A182XEG8 A0A182MQR2 A0A182WM19 A0A1I8N4T2 A0A182RE50 A0A182TD49 A0A182Q6R4 A0A182N1Y9 T1PC05 A0A1B0C6Q0 A0A1A9ZDX0 A0A146MFQ9 A0A1A9WLL5 A0A084WE14 A0A0P6IY33 A0A034WPS5 A0A0K8TVX2 A0A0A1XQY2 A0A1A9XJV5 A0A1A9UKQ9 A0A182H2Z8 W8BBQ2 A0A2R5LEC8 A0A1B0GF73 W5JV58 A0A2M4AKP4 K1RS36 A0A2M4AJT4 A0A1Q3G2N0 A0A210QT24 A0A1S3JDM1 A0A224YK68 A0A131YJI1 A0A131YE58 A0A1J1I5K2 L7MGY1
Pubmed
19121390
23622113
22118469
26354079
26227816
18362917
+ More
19820115 24845553 20798317 28004739 21282665 21719571 21347285 20566863 24508170 30249741 23691247 26334808 25401762 20966253 12364791 14747013 17210077 28648823 26108605 27129103 25474469 25244985 25315136 26823975 24438588 26999592 25348373 25830018 26483478 24495485 20920257 23761445 22992520 28812685 26383154 28797301 26830274 25576852
19820115 24845553 20798317 28004739 21282665 21719571 21347285 20566863 24508170 30249741 23691247 26334808 25401762 20966253 12364791 14747013 17210077 28648823 26108605 27129103 25474469 25244985 25315136 26823975 24438588 26999592 25348373 25830018 26483478 24495485 20920257 23761445 22992520 28812685 26383154 28797301 26830274 25576852
EMBL
BABH01011964
DQ311361
ABD36305.1
NWSH01000114
PCG79410.1
ODYU01006722
+ More
SOQ48823.1 GAIX01000113 JAA92447.1 RSAL01000416 RVE41814.1 AGBW02008299 OWR53794.1 KQ460205 KPJ16800.1 JTDY01001185 KOB74616.1 KQ971319 EFA00464.1 NEVH01011228 PNF31628.1 LJIG01016106 KRT81598.1 PNF31629.1 KK852567 KDR21197.1 GL451188 EFN79800.1 GEZM01061168 JAV70714.1 GEZM01061169 JAV70713.1 GFDF01003384 JAV10700.1 GL437365 EFN70555.1 GL761846 EFZ22741.1 KQ977642 KYN01042.1 KQ981954 KYN32175.1 LBMM01004181 KMQ92724.1 KQ976600 KYM79473.1 KZ288193 PBC34049.1 KQ980629 KYN14995.1 GL888217 EGI64602.1 GEBQ01012928 JAT27049.1 ADTU01010271 KQ982519 KYQ55486.1 AJVK01013756 AJVK01013757 AAZO01005714 DS235815 EEB17531.1 KQ414875 KOC59916.1 KK107231 QOIP01000009 EZA54973.1 RLU18601.1 KQ435007 KZC13519.1 JH431785 KK854311 PTY14506.1 KQ759770 OAD62933.1 AK418112 BAN21327.1 GECZ01029683 JAS40086.1 GEDC01004813 JAS32485.1 AJWK01035499 AJWK01035500 AJWK01035501 GECL01001420 JAP04704.1 GBGD01001306 JAC87583.1 GFTR01006572 JAW09854.1 GBHO01029045 JAG14559.1 GBRD01011480 GBRD01011479 JAG54345.1 AAAB01008948 EAA10359.4 APCN01004655 NNAY01000462 OXU28202.1 KK121203 KFM79956.1 JRES01000760 KNC28503.1 GDKW01001876 JAI54719.1 ACPB03001908 GAHY01001625 JAA75885.1 GBBI01000732 JAC17980.1 AXCM01003001 AXCN02000977 KA646302 AFP60931.1 JXJN01026846 GDHC01000001 JAQ18628.1 ATLV01023101 KE525340 KFB48458.1 GDUN01000213 JAN95706.1 GAKP01003219 GAKP01003217 JAC55733.1 GDHF01033690 JAI18624.1 GBXI01001309 JAD12983.1 JXUM01024326 KQ560687 KXJ81265.1 GAMC01016001 GAMC01015999 JAB90556.1 GGLE01003682 MBY07808.1 CCAG010012769 ADMH02000266 ETN67188.1 GGFK01008032 MBW41353.1 JH816099 EKC37296.1 GGFK01007722 MBW41043.1 GFDL01001046 JAV33999.1 NEDP02002059 OWF51862.1 GFPF01006901 MAA18047.1 GEDV01009829 JAP78728.1 GEDV01012366 JAP76191.1 CVRI01000042 CRK95560.1 GACK01001982 JAA63052.1
SOQ48823.1 GAIX01000113 JAA92447.1 RSAL01000416 RVE41814.1 AGBW02008299 OWR53794.1 KQ460205 KPJ16800.1 JTDY01001185 KOB74616.1 KQ971319 EFA00464.1 NEVH01011228 PNF31628.1 LJIG01016106 KRT81598.1 PNF31629.1 KK852567 KDR21197.1 GL451188 EFN79800.1 GEZM01061168 JAV70714.1 GEZM01061169 JAV70713.1 GFDF01003384 JAV10700.1 GL437365 EFN70555.1 GL761846 EFZ22741.1 KQ977642 KYN01042.1 KQ981954 KYN32175.1 LBMM01004181 KMQ92724.1 KQ976600 KYM79473.1 KZ288193 PBC34049.1 KQ980629 KYN14995.1 GL888217 EGI64602.1 GEBQ01012928 JAT27049.1 ADTU01010271 KQ982519 KYQ55486.1 AJVK01013756 AJVK01013757 AAZO01005714 DS235815 EEB17531.1 KQ414875 KOC59916.1 KK107231 QOIP01000009 EZA54973.1 RLU18601.1 KQ435007 KZC13519.1 JH431785 KK854311 PTY14506.1 KQ759770 OAD62933.1 AK418112 BAN21327.1 GECZ01029683 JAS40086.1 GEDC01004813 JAS32485.1 AJWK01035499 AJWK01035500 AJWK01035501 GECL01001420 JAP04704.1 GBGD01001306 JAC87583.1 GFTR01006572 JAW09854.1 GBHO01029045 JAG14559.1 GBRD01011480 GBRD01011479 JAG54345.1 AAAB01008948 EAA10359.4 APCN01004655 NNAY01000462 OXU28202.1 KK121203 KFM79956.1 JRES01000760 KNC28503.1 GDKW01001876 JAI54719.1 ACPB03001908 GAHY01001625 JAA75885.1 GBBI01000732 JAC17980.1 AXCM01003001 AXCN02000977 KA646302 AFP60931.1 JXJN01026846 GDHC01000001 JAQ18628.1 ATLV01023101 KE525340 KFB48458.1 GDUN01000213 JAN95706.1 GAKP01003219 GAKP01003217 JAC55733.1 GDHF01033690 JAI18624.1 GBXI01001309 JAD12983.1 JXUM01024326 KQ560687 KXJ81265.1 GAMC01016001 GAMC01015999 JAB90556.1 GGLE01003682 MBY07808.1 CCAG010012769 ADMH02000266 ETN67188.1 GGFK01008032 MBW41353.1 JH816099 EKC37296.1 GGFK01007722 MBW41043.1 GFDL01001046 JAV33999.1 NEDP02002059 OWF51862.1 GFPF01006901 MAA18047.1 GEDV01009829 JAP78728.1 GEDV01012366 JAP76191.1 CVRI01000042 CRK95560.1 GACK01001982 JAA63052.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000037510
+ More
UP000007266 UP000235965 UP000027135 UP000008237 UP000000311 UP000078542 UP000078541 UP000036403 UP000078540 UP000242457 UP000005203 UP000078492 UP000007755 UP000005205 UP000075809 UP000092462 UP000009046 UP000053825 UP000053097 UP000279307 UP000192223 UP000076502 UP000092461 UP000075881 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000215335 UP000075880 UP000054359 UP000037069 UP000095300 UP000075901 UP000015103 UP000076408 UP000076407 UP000075883 UP000075920 UP000095301 UP000075900 UP000075902 UP000075886 UP000075884 UP000092460 UP000092445 UP000091820 UP000030765 UP000092443 UP000078200 UP000069940 UP000249989 UP000092444 UP000000673 UP000005408 UP000242188 UP000085678 UP000183832
UP000007266 UP000235965 UP000027135 UP000008237 UP000000311 UP000078542 UP000078541 UP000036403 UP000078540 UP000242457 UP000005203 UP000078492 UP000007755 UP000005205 UP000075809 UP000092462 UP000009046 UP000053825 UP000053097 UP000279307 UP000192223 UP000076502 UP000092461 UP000075881 UP000075885 UP000075903 UP000075882 UP000007062 UP000075840 UP000215335 UP000075880 UP000054359 UP000037069 UP000095300 UP000075901 UP000015103 UP000076408 UP000076407 UP000075883 UP000075920 UP000095301 UP000075900 UP000075902 UP000075886 UP000075884 UP000092460 UP000092445 UP000091820 UP000030765 UP000092443 UP000078200 UP000069940 UP000249989 UP000092444 UP000000673 UP000005408 UP000242188 UP000085678 UP000183832
PRIDE
Pfam
Interpro
IPR033008
CSN1
+ More
IPR036390 WH_DNA-bd_sf
IPR000717 PCI_dom
IPR019585 Rpn7/CSN1
IPR009057 Homeobox-like_sf
IPR001356 Homeobox_dom
IPR010482 Peroxin
IPR006614 Peroxin/Ferlin
IPR006624 Beta-propeller_rpt_TECPR
IPR011949 HAD-SF_hydro_IA_REG-2-like
IPR023214 HAD_sf
IPR041492 HAD_2
IPR036412 HAD-like_sf
IPR006439 HAD-SF_hydro_IA
IPR011990 TPR-like_helical_dom_sf
IPR036390 WH_DNA-bd_sf
IPR000717 PCI_dom
IPR019585 Rpn7/CSN1
IPR009057 Homeobox-like_sf
IPR001356 Homeobox_dom
IPR010482 Peroxin
IPR006614 Peroxin/Ferlin
IPR006624 Beta-propeller_rpt_TECPR
IPR011949 HAD-SF_hydro_IA_REG-2-like
IPR023214 HAD_sf
IPR041492 HAD_2
IPR036412 HAD-like_sf
IPR006439 HAD-SF_hydro_IA
IPR011990 TPR-like_helical_dom_sf
Gene 3D
ProteinModelPortal
Q2F5R2
A0A2A4K6X6
A0A2H1W709
S4Q0A8
A0A3S2LQQ0
A0A212FJ83
+ More
A0A194RHK1 A0A0L7LHE4 D6WF13 A0A2J7QSS0 A0A0T6B394 A0A2J7QSR7 A0A067RBT3 E2BWT6 A0A1Y1LAQ0 A0A1Y1LAP6 A0A1L8DW78 E2A7N4 E9I9P5 A0A195CKR1 A0A195EV85 A0A0J7KR61 A0A151I0R2 A0A2A3ESE3 A0A088AP80 A0A151J0G6 F4WMJ1 A0A1B6LTV4 A0A158NAE9 A0A151X4Y1 A0A1B0DBQ5 E0VVX5 A0A0L7QN62 A0A026WG97 A0A1W4WN96 A0A154PNT6 T1J1H8 A0A2R7W465 A0A310STA3 R4WTT0 A0A1B6EQ85 A0A1B6E3J4 A0A1B0CY66 A0A0V0G9E2 A0A069DTZ7 A0A224XFP5 A0A0A9X3K0 A0A182JVZ8 A0A182PTV4 A0A0K8SNE6 A0A182UWY4 A0A182KNP6 Q7Q7Y5 A0A182HQL1 A0A232FC43 A0A182J1J0 A0A087URG7 A0A0L0C7W4 A0A0P4VQI5 A0A1I8NY35 A0A182SKX2 R4G461 A0A023F9C2 A0A182YR54 A0A182XEG8 A0A182MQR2 A0A182WM19 A0A1I8N4T2 A0A182RE50 A0A182TD49 A0A182Q6R4 A0A182N1Y9 T1PC05 A0A1B0C6Q0 A0A1A9ZDX0 A0A146MFQ9 A0A1A9WLL5 A0A084WE14 A0A0P6IY33 A0A034WPS5 A0A0K8TVX2 A0A0A1XQY2 A0A1A9XJV5 A0A1A9UKQ9 A0A182H2Z8 W8BBQ2 A0A2R5LEC8 A0A1B0GF73 W5JV58 A0A2M4AKP4 K1RS36 A0A2M4AJT4 A0A1Q3G2N0 A0A210QT24 A0A1S3JDM1 A0A224YK68 A0A131YJI1 A0A131YE58 A0A1J1I5K2 L7MGY1
A0A194RHK1 A0A0L7LHE4 D6WF13 A0A2J7QSS0 A0A0T6B394 A0A2J7QSR7 A0A067RBT3 E2BWT6 A0A1Y1LAQ0 A0A1Y1LAP6 A0A1L8DW78 E2A7N4 E9I9P5 A0A195CKR1 A0A195EV85 A0A0J7KR61 A0A151I0R2 A0A2A3ESE3 A0A088AP80 A0A151J0G6 F4WMJ1 A0A1B6LTV4 A0A158NAE9 A0A151X4Y1 A0A1B0DBQ5 E0VVX5 A0A0L7QN62 A0A026WG97 A0A1W4WN96 A0A154PNT6 T1J1H8 A0A2R7W465 A0A310STA3 R4WTT0 A0A1B6EQ85 A0A1B6E3J4 A0A1B0CY66 A0A0V0G9E2 A0A069DTZ7 A0A224XFP5 A0A0A9X3K0 A0A182JVZ8 A0A182PTV4 A0A0K8SNE6 A0A182UWY4 A0A182KNP6 Q7Q7Y5 A0A182HQL1 A0A232FC43 A0A182J1J0 A0A087URG7 A0A0L0C7W4 A0A0P4VQI5 A0A1I8NY35 A0A182SKX2 R4G461 A0A023F9C2 A0A182YR54 A0A182XEG8 A0A182MQR2 A0A182WM19 A0A1I8N4T2 A0A182RE50 A0A182TD49 A0A182Q6R4 A0A182N1Y9 T1PC05 A0A1B0C6Q0 A0A1A9ZDX0 A0A146MFQ9 A0A1A9WLL5 A0A084WE14 A0A0P6IY33 A0A034WPS5 A0A0K8TVX2 A0A0A1XQY2 A0A1A9XJV5 A0A1A9UKQ9 A0A182H2Z8 W8BBQ2 A0A2R5LEC8 A0A1B0GF73 W5JV58 A0A2M4AKP4 K1RS36 A0A2M4AJT4 A0A1Q3G2N0 A0A210QT24 A0A1S3JDM1 A0A224YK68 A0A131YJI1 A0A131YE58 A0A1J1I5K2 L7MGY1
PDB
4WSN
E-value=0,
Score=1639
Ontologies
GO
PANTHER
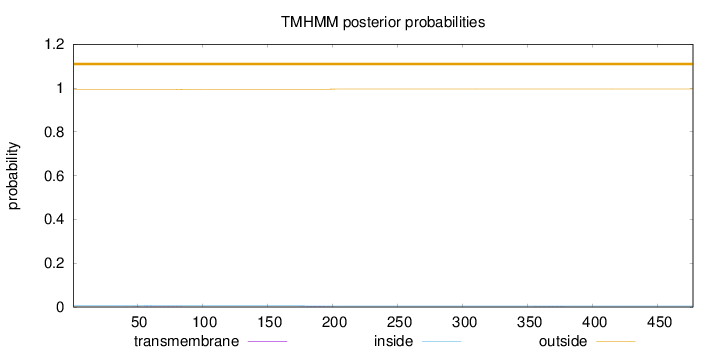
Topology
Subcellular location
Nucleus
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06345
Exp number, first 60 AAs:
0.00431
Total prob of N-in:
0.00665
outside
1 - 477
Population Genetic Test Statistics
Pi
48.852117
Theta
156.750765
Tajima's D
-2.195837
CLR
2222.558656
CSRT
0.00619969001549923
Interpretation
Uncertain
