Pre Gene Modal
BGIBMGA011711
Annotation
PREDICTED:_DIS3-like_exonuclease_2_[Amyelois_transitella]
Full name
DIS3-like exonuclease 2
Location in the cell
Cytoplasmic Reliability : 2.152 Nuclear Reliability : 1.72
Sequence
CDS
ATGAGAGATATTTTACAAAGCCCAGAGCATGTTGCTAGTGTACCTATCACTCCATTAATATTATCAGGACAGAGATCAGAACCTGTTTGTATTCCAAATTCTACATTTATTCATTCTGAACCTAGCACTCCAAGGTTCATAGAGAATGCATTGCAACACCCCTTATTGAATCCATCATTTTCAAGACAAACTGGTACAATACAAATTGGACAGCTCAGACTGAGGCATAATTCAGAAAACAATTGCTTAACTAAAACTCTTGTAAATGGTGACAAGAAATTTGATGCATACCTTTCTGGAGAAGATGTTGAGAAAGGTTTAAAGAACAATATGCTTATTGAAGGTGTTCTGAGGATAAATCCAAAACAATATCAACATTCATATGTTTCAAGTCAAGACCGGAGTCAACAAGATATATTAATAGACGGTATTAAGAATAGAAATCGTGCAATGGAAGGAGATGTAGTAATAGTACAATTAGTTGAAAATGAACCTGAAGAAAACGTTACTAATCCTGAACAAAAAAGAGGGAAAGTAGTTTTTATCAAGGAAAAAGTTCATACTAGAACTTGTATTGGTTCACTCAAACTAATGCATGACAAAAATAGACAAAAAGCTATGTTCGTGCCTCGAGATCACAGAGTACCAAGAATAAATATACCATTTACTTCATGGCCAGATAATTTTTATGAAGATGCCAAGAAATATGAAAACACATTGTTTGTTGCAAAAATAATAGATTGGTCTGATACAAGGTTTGCTGTAGGAAAAATTATGGGTAATATTGGTAAATCAGGAGACATGGTCACTGAAACAAAAGCAATATTAGCAGAAAATGATTTAGATGTTACTGCTTTTGGCTCTGAATACAAGGATTTGTATCCTCAACTTGACTTTACAATTCCCGATGAAGAAATAGCAAAGAGAGAAGACTGTCGGATGTTATGTATCTTTAGTATTGATCCATTTAATTGCCGTGATATTGATGATGCAGTCTCTTGCAAGGAACTAGAAAATGGTAATTATGAGATCGGAGTCCATATTTCTGATGTTACATATTTTTTAAATGAAAACACTCCGCTTGATGTGAAAGTTGCAGAAAAAGCTACAACAATATATTTGGTAGAAACTGCTTACCACATGCTTCCAGATGATTTGTGTATGCTTTGTTCTCTATTCCCTGGTGTAGATAAACTAGCTTTCTCTGTTTTTTGGGAAATGACCAAGGATGCTAAAGTTATTAGTTACAGGTTTGCTAGGACTGTTATAAATTCATGCTGTCAGTTGGCATATGAACATGCTCAAGATGTTCTTGATGACAAAGAGGATGCTCAAATAAAATTTCCAGAAATGTACAATGGTTTTGATTACAAAGATGTGCACAAATCAATAAAAATTTTAGGCAAATTGTCAAATATTCTTCGTGCAAATAGATTTGAGAATGGTGCATTGAGAATTGATCAACCTAAGGCTAGTTTTAGATTAAATCCTACTAGTGGTTTACCAGAGTCATACTGCATATATGAGAGTAAAGAAAGCCATCAACTCATTGAGGAATTCATGTTGCTCGCTAACATGTCTGTAGCTACACGTATTTATAGTGACCACCCAAAACTGGCATTTTTAAGGAGTCATCCACCACCAAGTAAATATATGTTGATGCAGCTAGAAAAGTCTCTAAAACCAATGGGCATAAAATTAGATATCGAATCGGCTGGACATCTACACAGATCTTTACTACCATATGTGGGACCAGACAATACAGACAAAGGAAGAGCCATGGTTTTAAATTTGCTATGTGCAAAACCTATGACAAGAGCAAAATATTTTTGTGCTGCTGCTTGTGATGATGATGAATTTCAACATTATGCTCTGAATGTGCCCCTGTACACTCATTTCACAAGTCCTATTAGGAGATATGCTGACATTATGGTTCATAGGCTATTAGCAGCAAGCTTAAATTATAGAGAAACTCCAGCATGGCAGGAAGATGATGTACGCATGATAGCTGCTCAATGTAACAAACAGAAGTACAATGCTAAAAAGGCTGGTGAAATGTCAACAGAATTGTACACTTTAAAGTACATTGAACTTAATTCTCCCATAGTCACTGAAGGTGCTGTGGTTGACATTAGGGAGAAATATATTGATGTTATCATTGTTTCCATGGGCCTGAACCGGAGAATATTTTTCAATAATGATTTTCCTGGAGAGTATAAGTGTATTAGACACGAAGAACTTGCGAAGTTGGCTCGAATGGAACTGAAATGGAAACCGGTGAATGAATTGCCAGAAACTAAACAAGTAATTGGAGTTTTTTCCATTGTACAAGTTGAAATGCACAAAGGCGATGACATGGATTACTATTTACAATTCATAGAACTGTCTTAA
Protein
MRDILQSPEHVASVPITPLILSGQRSEPVCIPNSTFIHSEPSTPRFIENALQHPLLNPSFSRQTGTIQIGQLRLRHNSENNCLTKTLVNGDKKFDAYLSGEDVEKGLKNNMLIEGVLRINPKQYQHSYVSSQDRSQQDILIDGIKNRNRAMEGDVVIVQLVENEPEENVTNPEQKRGKVVFIKEKVHTRTCIGSLKLMHDKNRQKAMFVPRDHRVPRINIPFTSWPDNFYEDAKKYENTLFVAKIIDWSDTRFAVGKIMGNIGKSGDMVTETKAILAENDLDVTAFGSEYKDLYPQLDFTIPDEEIAKREDCRMLCIFSIDPFNCRDIDDAVSCKELENGNYEIGVHISDVTYFLNENTPLDVKVAEKATTIYLVETAYHMLPDDLCMLCSLFPGVDKLAFSVFWEMTKDAKVISYRFARTVINSCCQLAYEHAQDVLDDKEDAQIKFPEMYNGFDYKDVHKSIKILGKLSNILRANRFENGALRIDQPKASFRLNPTSGLPESYCIYESKESHQLIEEFMLLANMSVATRIYSDHPKLAFLRSHPPPSKYMLMQLEKSLKPMGIKLDIESAGHLHRSLLPYVGPDNTDKGRAMVLNLLCAKPMTRAKYFCAAACDDDEFQHYALNVPLYTHFTSPIRRYADIMVHRLLAASLNYRETPAWQEDDVRMIAAQCNKQKYNAKKAGEMSTELYTLKYIELNSPIVTEGAVVDIREKYIDVIIVSMGLNRRIFFNNDFPGEYKCIRHEELAKLARMELKWKPVNELPETKQVIGVFSIVQVEMHKGDDMDYYLQFIELS
Summary
Description
3'-5'-exoribonuclease that specifically recognizes RNAs polyuridylated at their 3' end and mediates their degradation. Component of an exosome-independent RNA degradation pathway that mediates degradation of both mRNAs and miRNAs that have been polyuridylated by a terminal uridylyltransferase. Essential for correct mitosis, and negatively regulates cell proliferation.
3'-5'-exoribonuclease that specifically recognizes RNAs polyuridylated at their 3' end and mediates their degradation. Component of an exosome-independent RNA degradation pathway that mediates degradation of both mRNAs and miRNAs that have been polyuridylated by a terminal uridylyltransferase, such as ZCCHC11/TUT4. Mediates degradation of cytoplasmic mRNAs that have been deadenylated and subsequently uridylated at their 3'. Mediates degradation of uridylated pre-let-7 miRNAs, contributing to the maintenance of embryonic stem (ES) cells. Essential for correct mitosis, and negatively regulates cell proliferation.
3'-5'-exoribonuclease that specifically recognizes RNAs polyuridylated at their 3' end and mediates their degradation. Component of an exosome-independent RNA degradation pathway that mediates degradation of cytoplasmic mRNAs that have been deadenylated and subsequently uridylated at their 3'.
3'-5'-exoribonuclease that specifically recognizes RNAs polyuridylated at their 3' end and mediates their degradation. Component of an exosome-independent RNA degradation pathway that mediates degradation of both mRNAs and miRNAs that have been polyuridylated by a terminal uridylyltransferase, such as ZCCHC11/TUT4. Mediates degradation of cytoplasmic mRNAs that have been deadenylated and subsequently uridylated at their 3'. Mediates degradation of uridylated pre-let-7 miRNAs, contributing to the maintenance of embryonic stem (ES) cells. Essential for correct mitosis, and negatively regulates cell proliferation.
3'-5'-exoribonuclease that specifically recognizes RNAs polyuridylated at their 3' end and mediates their degradation. Component of an exosome-independent RNA degradation pathway that mediates degradation of cytoplasmic mRNAs that have been deadenylated and subsequently uridylated at their 3'.
Subunit
Interacts with XRN1.
Similarity
Belongs to the RNR ribonuclease family.
Belongs to the RNR ribonuclease family. DIS3L2 subfamily.
Belongs to the RNR ribonuclease family. DIS3L2 subfamily.
Keywords
Acetylation
Alternative splicing
Cell cycle
Cell division
Complete proteome
Cytoplasm
Disease mutation
Exonuclease
Hydrolase
Magnesium
Metal-binding
Mitosis
Nuclease
Phosphoprotein
Polymorphism
Reference proteome
RNA-binding
Tumor suppressor
3D-structure
Feature
chain DIS3-like exonuclease 2
splice variant In isoform 5.
sequence variant In dbSNP:rs723044.
splice variant In isoform 5.
sequence variant In dbSNP:rs723044.
Uniprot
A0A2A4K670
A0A194QP61
A0A194PVQ3
A0A2H1W8L0
A0A0L7LCQ7
A0A067RGS9
+ More
A0A1W4WUX9 A0A1W4WV58 A0A139WCX3 A0A1B6GGU6 A0A1B6KK62 E0VYX1 A0A0K8THG4 A0A146LQG2 A0A0A9VVG8 A0A087ZSA6 A0A224XK04 A0A0V0GAH1 A0A0L7RD70 A0A2A3EN79 A0A1Y1MUI4 A0A3Q2YWN3 A0A2R8Q8H5 T1I3N0 A0A336L9E5 A0A336M229 A0A154PJ60 A0A3B5QPE4 A0A3B3VYR2 A0A3Q7T152 K7ISD2 A0A3B4B956 E7F350 A0A3Q3DQN5 F6XKS7 M3WH44 A0A3Q7U436 A0A340XRJ3 A0A341B3G2 A0A3Q0REW6 A0A3Q3N439 A0A384ABQ1 H0WZY7 A0A2U3VM61 A0A3Q7QHJ6 A0A2Y9N329 A0A3Q2GXI4 F6U4I9 A0A384ABR4 L5M1S6 A0A3Q2H0F0 A0A2Y9H352 L8HZU2 A0A384C1Q5 D2H854 E2BZ80 G1MDU8 I3M6I1 W5QHT9 A0A3Q7VY81 A0A250Y5W0 E1B7R0 A0A2K6TCX9 A0A2K6GM68 I3LMZ3 D3K5M4 A0A3P4N016 A0A182JQZ8 M3Y0K2 A0A2Y9FKK5 Q8IYB7 L5L0E3 A0A232F777 U3BGP9 H2R1E6 A0A2R9ALF7 C3XYF0 T2M9C5 A0A2I4ARZ8 V4B8T0 A0A2K5C716 A0A2J8TQB3 A0A341B0N4 F7C0G6 F4X1K7 A0A2Y9J0I4 A0A3Q7QHL0 K9IN83 A0A3B4DFG4 A0A2K6ABW3 A0A2K6QIP8 Q8CI75 Q8CI75-2 A0A2Y9MXB0 A0A1U7R708 H2M031 H9FPW8
A0A1W4WUX9 A0A1W4WV58 A0A139WCX3 A0A1B6GGU6 A0A1B6KK62 E0VYX1 A0A0K8THG4 A0A146LQG2 A0A0A9VVG8 A0A087ZSA6 A0A224XK04 A0A0V0GAH1 A0A0L7RD70 A0A2A3EN79 A0A1Y1MUI4 A0A3Q2YWN3 A0A2R8Q8H5 T1I3N0 A0A336L9E5 A0A336M229 A0A154PJ60 A0A3B5QPE4 A0A3B3VYR2 A0A3Q7T152 K7ISD2 A0A3B4B956 E7F350 A0A3Q3DQN5 F6XKS7 M3WH44 A0A3Q7U436 A0A340XRJ3 A0A341B3G2 A0A3Q0REW6 A0A3Q3N439 A0A384ABQ1 H0WZY7 A0A2U3VM61 A0A3Q7QHJ6 A0A2Y9N329 A0A3Q2GXI4 F6U4I9 A0A384ABR4 L5M1S6 A0A3Q2H0F0 A0A2Y9H352 L8HZU2 A0A384C1Q5 D2H854 E2BZ80 G1MDU8 I3M6I1 W5QHT9 A0A3Q7VY81 A0A250Y5W0 E1B7R0 A0A2K6TCX9 A0A2K6GM68 I3LMZ3 D3K5M4 A0A3P4N016 A0A182JQZ8 M3Y0K2 A0A2Y9FKK5 Q8IYB7 L5L0E3 A0A232F777 U3BGP9 H2R1E6 A0A2R9ALF7 C3XYF0 T2M9C5 A0A2I4ARZ8 V4B8T0 A0A2K5C716 A0A2J8TQB3 A0A341B0N4 F7C0G6 F4X1K7 A0A2Y9J0I4 A0A3Q7QHL0 K9IN83 A0A3B4DFG4 A0A2K6ABW3 A0A2K6QIP8 Q8CI75 Q8CI75-2 A0A2Y9MXB0 A0A1U7R708 H2M031 H9FPW8
EC Number
3.1.13.-
Pubmed
26354079
26227816
24845553
18362917
19820115
20566863
+ More
26823975 25401762 28004739 23594743 23542700 20075255 25463417 16341006 17975172 19892987 22751099 20010809 20798317 20809919 28087693 19393038 30723633 14702039 17974005 15815621 15489334 11352565 17373680 19608861 20068231 21269460 22306653 23613427 23756462 23486540 23186163 24141620 24275569 28328139 23258410 28648823 25243066 16136131 22722832 18563158 24065732 23254933 21719571 25362486 16141072 17242355 21183079 23594738 25119025 17554307 25319552
26823975 25401762 28004739 23594743 23542700 20075255 25463417 16341006 17975172 19892987 22751099 20010809 20798317 20809919 28087693 19393038 30723633 14702039 17974005 15815621 15489334 11352565 17373680 19608861 20068231 21269460 22306653 23613427 23756462 23486540 23186163 24141620 24275569 28328139 23258410 28648823 25243066 16136131 22722832 18563158 24065732 23254933 21719571 25362486 16141072 17242355 21183079 23594738 25119025 17554307 25319552
EMBL
NWSH01000114
PCG79418.1
KQ461190
KPJ07139.1
KQ459590
KPI97407.1
+ More
ODYU01006722 SOQ48824.1 JTDY01001634 KOB73283.1 KK852649 KDR19486.1 KQ971361 KYB25799.1 GECZ01008256 JAS61513.1 GEBQ01028135 JAT11842.1 DS235848 EEB18577.1 GBRD01000862 JAG64959.1 GDHC01009627 JAQ09002.1 GBHO01043935 JAF99668.1 GFTR01008065 JAW08361.1 GECL01001879 JAP04245.1 KQ414615 KOC68745.1 KZ288204 PBC33188.1 GEZM01020180 JAV89332.1 BX001029 ACPB03027837 UFQS01002148 UFQT01002148 SSX13333.1 SSX32767.1 UFQT01000305 SSX23059.1 KQ434931 KZC11857.1 AAEX03014441 AAEX03014442 AAEX03014443 AAEX03014444 AAEX03014445 AANG04002752 AAQR03029964 AAQR03029965 AAQR03029966 AAQR03029967 AAQR03029968 AAQR03029969 AAQR03029970 AAQR03029971 AAQR03029972 AAQR03029973 KB105166 ELK32599.1 JH882669 ELR48582.1 GL192569 EFB20663.1 GL451577 EFN78986.1 ACTA01040620 ACTA01048620 ACTA01056620 ACTA01064620 ACTA01072620 ACTA01080620 ACTA01088620 ACTA01096620 ACTA01104620 ACTA01112620 AGTP01044221 AGTP01044222 AGTP01044223 AGTP01044224 AGTP01044225 AGTP01044226 AGTP01044227 AMGL01056635 AMGL01056636 AMGL01056637 AMGL01056638 AMGL01056639 AMGL01056640 AMGL01056641 AMGL01056642 AMGL01056643 GFFV01001011 JAV38934.1 AEMK02000101 DQIR01188270 DQIR01296152 HDB43747.1 GU373702 ADC38902.1 CYRY02015647 VCW85551.1 AEYP01081372 AEYP01081373 AEYP01081374 AEYP01081375 AEYP01081376 AEYP01081377 AEYP01081378 AEYP01081379 AEYP01081380 AEYP01081381 AK094293 AL834174 BX648325 AF443854 AC019130 AC068134 AC093374 AC138658 BC026166 BC036113 KB030407 ELK17107.1 NNAY01000827 OXU26298.1 GAMT01006972 GAMR01010171 JAB04889.1 JAB23761.1 AACZ04056640 AC195510 GABC01003460 GABF01002970 GABD01001901 GABE01003523 NBAG03000230 JAA07878.1 JAA19175.1 JAA31199.1 JAA41216.1 PNI70347.1 AJFE02075488 AJFE02075489 AJFE02075490 AJFE02075491 AJFE02075492 AJFE02075493 AJFE02075494 AJFE02075495 AJFE02075496 GG666473 EEN66835.1 HAAD01002671 CDG68903.1 KB203301 ESO85209.1 NDHI03003487 PNJ35173.1 GAMS01007378 GAMQ01005731 GAMP01004324 JAB15758.1 JAB36120.1 JAB48431.1 GL888531 EGI59665.1 GABZ01004252 JAA49273.1 AK031180 BC036177 JU332922 AFE76677.1
ODYU01006722 SOQ48824.1 JTDY01001634 KOB73283.1 KK852649 KDR19486.1 KQ971361 KYB25799.1 GECZ01008256 JAS61513.1 GEBQ01028135 JAT11842.1 DS235848 EEB18577.1 GBRD01000862 JAG64959.1 GDHC01009627 JAQ09002.1 GBHO01043935 JAF99668.1 GFTR01008065 JAW08361.1 GECL01001879 JAP04245.1 KQ414615 KOC68745.1 KZ288204 PBC33188.1 GEZM01020180 JAV89332.1 BX001029 ACPB03027837 UFQS01002148 UFQT01002148 SSX13333.1 SSX32767.1 UFQT01000305 SSX23059.1 KQ434931 KZC11857.1 AAEX03014441 AAEX03014442 AAEX03014443 AAEX03014444 AAEX03014445 AANG04002752 AAQR03029964 AAQR03029965 AAQR03029966 AAQR03029967 AAQR03029968 AAQR03029969 AAQR03029970 AAQR03029971 AAQR03029972 AAQR03029973 KB105166 ELK32599.1 JH882669 ELR48582.1 GL192569 EFB20663.1 GL451577 EFN78986.1 ACTA01040620 ACTA01048620 ACTA01056620 ACTA01064620 ACTA01072620 ACTA01080620 ACTA01088620 ACTA01096620 ACTA01104620 ACTA01112620 AGTP01044221 AGTP01044222 AGTP01044223 AGTP01044224 AGTP01044225 AGTP01044226 AGTP01044227 AMGL01056635 AMGL01056636 AMGL01056637 AMGL01056638 AMGL01056639 AMGL01056640 AMGL01056641 AMGL01056642 AMGL01056643 GFFV01001011 JAV38934.1 AEMK02000101 DQIR01188270 DQIR01296152 HDB43747.1 GU373702 ADC38902.1 CYRY02015647 VCW85551.1 AEYP01081372 AEYP01081373 AEYP01081374 AEYP01081375 AEYP01081376 AEYP01081377 AEYP01081378 AEYP01081379 AEYP01081380 AEYP01081381 AK094293 AL834174 BX648325 AF443854 AC019130 AC068134 AC093374 AC138658 BC026166 BC036113 KB030407 ELK17107.1 NNAY01000827 OXU26298.1 GAMT01006972 GAMR01010171 JAB04889.1 JAB23761.1 AACZ04056640 AC195510 GABC01003460 GABF01002970 GABD01001901 GABE01003523 NBAG03000230 JAA07878.1 JAA19175.1 JAA31199.1 JAA41216.1 PNI70347.1 AJFE02075488 AJFE02075489 AJFE02075490 AJFE02075491 AJFE02075492 AJFE02075493 AJFE02075494 AJFE02075495 AJFE02075496 GG666473 EEN66835.1 HAAD01002671 CDG68903.1 KB203301 ESO85209.1 NDHI03003487 PNJ35173.1 GAMS01007378 GAMQ01005731 GAMP01004324 JAB15758.1 JAB36120.1 JAB48431.1 GL888531 EGI59665.1 GABZ01004252 JAA49273.1 AK031180 BC036177 JU332922 AFE76677.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000037510
UP000027135
UP000192223
+ More
UP000007266 UP000009046 UP000005203 UP000053825 UP000242457 UP000264820 UP000000437 UP000015103 UP000076502 UP000002852 UP000261500 UP000286640 UP000002358 UP000261520 UP000002254 UP000011712 UP000265300 UP000252040 UP000261340 UP000261640 UP000261681 UP000005225 UP000245340 UP000286641 UP000248483 UP000002281 UP000248481 UP000261680 UP000008237 UP000008912 UP000005215 UP000002356 UP000286642 UP000009136 UP000233220 UP000233160 UP000008227 UP000075881 UP000000715 UP000248484 UP000005640 UP000010552 UP000215335 UP000002277 UP000240080 UP000001554 UP000192220 UP000030746 UP000233020 UP000007755 UP000248482 UP000261440 UP000233140 UP000233200 UP000000589 UP000189706 UP000001038
UP000007266 UP000009046 UP000005203 UP000053825 UP000242457 UP000264820 UP000000437 UP000015103 UP000076502 UP000002852 UP000261500 UP000286640 UP000002358 UP000261520 UP000002254 UP000011712 UP000265300 UP000252040 UP000261340 UP000261640 UP000261681 UP000005225 UP000245340 UP000286641 UP000248483 UP000002281 UP000248481 UP000261680 UP000008237 UP000008912 UP000005215 UP000002356 UP000286642 UP000009136 UP000233220 UP000233160 UP000008227 UP000075881 UP000000715 UP000248484 UP000005640 UP000010552 UP000215335 UP000002277 UP000240080 UP000001554 UP000192220 UP000030746 UP000233020 UP000007755 UP000248482 UP000261440 UP000233140 UP000233200 UP000000589 UP000189706 UP000001038
Pfam
Interpro
IPR033771
Rrp44_CSD1
+ More
IPR012340 NA-bd_OB-fold
IPR022966 RNase_II/R_CS
IPR001900 RNase_II/R
IPR041505 Dis3_CSD2
IPR041093 Dis3l2_C
IPR026850 FANCL_C
IPR019162 FancL_WD-rpt_cont_dom
IPR013083 Znf_RING/FYVE/PHD
IPR028591 DIS3L2
IPR019775 WD40_repeat_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR012340 NA-bd_OB-fold
IPR022966 RNase_II/R_CS
IPR001900 RNase_II/R
IPR041505 Dis3_CSD2
IPR041093 Dis3l2_C
IPR026850 FANCL_C
IPR019162 FancL_WD-rpt_cont_dom
IPR013083 Znf_RING/FYVE/PHD
IPR028591 DIS3L2
IPR019775 WD40_repeat_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
Gene 3D
ProteinModelPortal
A0A2A4K670
A0A194QP61
A0A194PVQ3
A0A2H1W8L0
A0A0L7LCQ7
A0A067RGS9
+ More
A0A1W4WUX9 A0A1W4WV58 A0A139WCX3 A0A1B6GGU6 A0A1B6KK62 E0VYX1 A0A0K8THG4 A0A146LQG2 A0A0A9VVG8 A0A087ZSA6 A0A224XK04 A0A0V0GAH1 A0A0L7RD70 A0A2A3EN79 A0A1Y1MUI4 A0A3Q2YWN3 A0A2R8Q8H5 T1I3N0 A0A336L9E5 A0A336M229 A0A154PJ60 A0A3B5QPE4 A0A3B3VYR2 A0A3Q7T152 K7ISD2 A0A3B4B956 E7F350 A0A3Q3DQN5 F6XKS7 M3WH44 A0A3Q7U436 A0A340XRJ3 A0A341B3G2 A0A3Q0REW6 A0A3Q3N439 A0A384ABQ1 H0WZY7 A0A2U3VM61 A0A3Q7QHJ6 A0A2Y9N329 A0A3Q2GXI4 F6U4I9 A0A384ABR4 L5M1S6 A0A3Q2H0F0 A0A2Y9H352 L8HZU2 A0A384C1Q5 D2H854 E2BZ80 G1MDU8 I3M6I1 W5QHT9 A0A3Q7VY81 A0A250Y5W0 E1B7R0 A0A2K6TCX9 A0A2K6GM68 I3LMZ3 D3K5M4 A0A3P4N016 A0A182JQZ8 M3Y0K2 A0A2Y9FKK5 Q8IYB7 L5L0E3 A0A232F777 U3BGP9 H2R1E6 A0A2R9ALF7 C3XYF0 T2M9C5 A0A2I4ARZ8 V4B8T0 A0A2K5C716 A0A2J8TQB3 A0A341B0N4 F7C0G6 F4X1K7 A0A2Y9J0I4 A0A3Q7QHL0 K9IN83 A0A3B4DFG4 A0A2K6ABW3 A0A2K6QIP8 Q8CI75 Q8CI75-2 A0A2Y9MXB0 A0A1U7R708 H2M031 H9FPW8
A0A1W4WUX9 A0A1W4WV58 A0A139WCX3 A0A1B6GGU6 A0A1B6KK62 E0VYX1 A0A0K8THG4 A0A146LQG2 A0A0A9VVG8 A0A087ZSA6 A0A224XK04 A0A0V0GAH1 A0A0L7RD70 A0A2A3EN79 A0A1Y1MUI4 A0A3Q2YWN3 A0A2R8Q8H5 T1I3N0 A0A336L9E5 A0A336M229 A0A154PJ60 A0A3B5QPE4 A0A3B3VYR2 A0A3Q7T152 K7ISD2 A0A3B4B956 E7F350 A0A3Q3DQN5 F6XKS7 M3WH44 A0A3Q7U436 A0A340XRJ3 A0A341B3G2 A0A3Q0REW6 A0A3Q3N439 A0A384ABQ1 H0WZY7 A0A2U3VM61 A0A3Q7QHJ6 A0A2Y9N329 A0A3Q2GXI4 F6U4I9 A0A384ABR4 L5M1S6 A0A3Q2H0F0 A0A2Y9H352 L8HZU2 A0A384C1Q5 D2H854 E2BZ80 G1MDU8 I3M6I1 W5QHT9 A0A3Q7VY81 A0A250Y5W0 E1B7R0 A0A2K6TCX9 A0A2K6GM68 I3LMZ3 D3K5M4 A0A3P4N016 A0A182JQZ8 M3Y0K2 A0A2Y9FKK5 Q8IYB7 L5L0E3 A0A232F777 U3BGP9 H2R1E6 A0A2R9ALF7 C3XYF0 T2M9C5 A0A2I4ARZ8 V4B8T0 A0A2K5C716 A0A2J8TQB3 A0A341B0N4 F7C0G6 F4X1K7 A0A2Y9J0I4 A0A3Q7QHL0 K9IN83 A0A3B4DFG4 A0A2K6ABW3 A0A2K6QIP8 Q8CI75 Q8CI75-2 A0A2Y9MXB0 A0A1U7R708 H2M031 H9FPW8
PDB
4PMW
E-value=1.68266e-140,
Score=1283
Ontologies
GO
Topology
Subcellular location
Cytoplasm
P-body
P-body
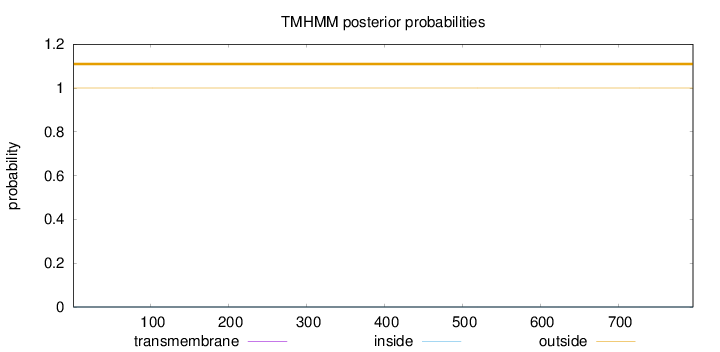
Length:
796
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000750000000000001
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00006
outside
1 - 796
Population Genetic Test Statistics
Pi
25.391663
Theta
55.353377
Tajima's D
-1.773859
CLR
2087.909899
CSRT
0.0299485025748713
Interpretation
Uncertain
