Gene
KWMTBOMO06661 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011771
Annotation
Uncharacterized_protein_OBRU01_10972_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 2.771
Sequence
CDS
ATGAAACTAATTACCCACAATATGCTCACTTCGAAATGCTTGAAAGGTGTTGTGACTGGCTATCCTCTTGCAATTAACGCGACAGACATTAAAGTTAAAGAAGTCGATTTTAATCCTGAATTTATTAGTCGTGTAATACCAAAGCTGGATTGGGAAGTATTATGGGTGGCCGCTGATAGCATCGGCCACAGTGATGGTCTGCCAAGATCTTTAGAAAATAAGTATGATGAAAACGAAGAGTTTTTGAAGAAAGCTCATAAAGTCTTGTTAGAGGTGGAAGTTGAAGAGGGACACTTGACTTGTCCAGAATCTGGAAGACAATTTCCGATTTCTAAAGGAATTCCTAATATGCTATTAAATGAAGCTGAAGTGCAGTAG
Protein
MKLITHNMLTSKCLKGVVTGYPLAINATDIKVKEVDFNPEFISRVIPKLDWEVLWVAADSIGHSDGLPRSLENKYDENEEFLKKAHKVLLEVEVEEGHLTCPESGRQFPISKGIPNMLLNEAEVQ
Summary
Uniprot
H9JQG5
A0A0L7LCR1
A0A2A4K5S7
A0A1E1WRK1
S4NUB3
A0A194RH80
+ More
A0A194PWM1 A0A212FJ82 A0A2H1W6U8 A0A1W4XGW0 A0A0L7RK31 A0A2A3EJ01 A0A088AQL9 N6TRU2 A0A1Y1NII6 A0A0P4W1G5 A0A0C9R5G8 A0A2J7QGF1 A0A023F4E5 A0A195F305 A0A2P8Z947 A0A151IG83 A0A195BCY4 F4WM83 E2BEA7 A0A1B6KY32 A0A0V0GDK3 A0A1B6H3A3 A0A310SNI7 A0A1B6JDB0 A0A1B6DIN9 A0A069DNL4 A0A151WFG5 A0A195DCD7 D6WHM6 A0A0N0BBK7 A0A0T6B2T4 A0A0A9WAC5 K7J0T0 T1I9K8 E2A0T1 A0A067R6E6 L7M605 A0A131YUI8 A0A131XCA8 A0A2R5LFV5 A0A0J7K7N4 E9FVZ3 A0A0C9SAE9 A0A224YYI2 A0A0P5LPI4 A0A023FWG6 A0A210QVU1 A0A1E1WZQ7 A0A1E1XU69 A0A023FDY7 A0A1S3HUW0 A0A026X3Q0 A0A158P2V8 A0A3Q3SWX1 A0A162SW91 A0A3Q2ZMR2 G3PXS1 A0A0P5XY68 A0A3Q1ITL6 A0A0P6GUF2 A0A3B3ZNH5 A0A3Q1BE39 A0A3P8SM81 A0A3Q4GDN3 A0A0K8RIC8 G3PXS4 A0A3Q1HS00 A0A3Q2Z8W9 A0A3Q2DU95 B7P923 A0A2U9C5T1 A0A0S7L872 A0A1A7WK78 A0A2I4B0M8 A0A3Q2VIW6 A0A3P8Q9J9 A0A3P9B990 A0A2I4B0P6 S4RJ54 C3KHA5 A0A1A8QF48 A0A1A8IBW6 A0A1A8GYF9 A0A3B3Y8G5 A0A3P9PL45 A0A315VFM9 A0A1A8DU99 A0A1A7ZP72 C3ZN06 A0A3B5QA98 A0A3B5KTH5 A0A0P4XD63 A0A3P8V317 H3BEP6
A0A194PWM1 A0A212FJ82 A0A2H1W6U8 A0A1W4XGW0 A0A0L7RK31 A0A2A3EJ01 A0A088AQL9 N6TRU2 A0A1Y1NII6 A0A0P4W1G5 A0A0C9R5G8 A0A2J7QGF1 A0A023F4E5 A0A195F305 A0A2P8Z947 A0A151IG83 A0A195BCY4 F4WM83 E2BEA7 A0A1B6KY32 A0A0V0GDK3 A0A1B6H3A3 A0A310SNI7 A0A1B6JDB0 A0A1B6DIN9 A0A069DNL4 A0A151WFG5 A0A195DCD7 D6WHM6 A0A0N0BBK7 A0A0T6B2T4 A0A0A9WAC5 K7J0T0 T1I9K8 E2A0T1 A0A067R6E6 L7M605 A0A131YUI8 A0A131XCA8 A0A2R5LFV5 A0A0J7K7N4 E9FVZ3 A0A0C9SAE9 A0A224YYI2 A0A0P5LPI4 A0A023FWG6 A0A210QVU1 A0A1E1WZQ7 A0A1E1XU69 A0A023FDY7 A0A1S3HUW0 A0A026X3Q0 A0A158P2V8 A0A3Q3SWX1 A0A162SW91 A0A3Q2ZMR2 G3PXS1 A0A0P5XY68 A0A3Q1ITL6 A0A0P6GUF2 A0A3B3ZNH5 A0A3Q1BE39 A0A3P8SM81 A0A3Q4GDN3 A0A0K8RIC8 G3PXS4 A0A3Q1HS00 A0A3Q2Z8W9 A0A3Q2DU95 B7P923 A0A2U9C5T1 A0A0S7L872 A0A1A7WK78 A0A2I4B0M8 A0A3Q2VIW6 A0A3P8Q9J9 A0A3P9B990 A0A2I4B0P6 S4RJ54 C3KHA5 A0A1A8QF48 A0A1A8IBW6 A0A1A8GYF9 A0A3B3Y8G5 A0A3P9PL45 A0A315VFM9 A0A1A8DU99 A0A1A7ZP72 C3ZN06 A0A3B5QA98 A0A3B5KTH5 A0A0P4XD63 A0A3P8V317 H3BEP6
Pubmed
19121390
26227816
23622113
26354079
22118469
23537049
+ More
28004739 27129103 25474469 29403074 21719571 20798317 26334808 18362917 19820115 25401762 20075255 24845553 25576852 26830274 28049606 21292972 26131772 28797301 28812685 28503490 29209593 24508170 30249741 21347285 25463417 25186727 29703783 18563158 23542700 24487278 9215903
28004739 27129103 25474469 29403074 21719571 20798317 26334808 18362917 19820115 25401762 20075255 24845553 25576852 26830274 28049606 21292972 26131772 28797301 28812685 28503490 29209593 24508170 30249741 21347285 25463417 25186727 29703783 18563158 23542700 24487278 9215903
EMBL
BABH01011965
JTDY01001634
KOB73288.1
NWSH01000114
PCG79419.1
GDQN01001598
+ More
JAT89456.1 GAIX01013377 JAA79183.1 KQ461190 KQ460205 KPJ07138.1 KPJ16799.1 KQ459590 KPI97408.1 AGBW02008299 OWR53791.1 ODYU01006722 SOQ48825.1 KQ414578 KOC71189.1 KZ288253 PBC31001.1 APGK01057714 KB741280 KB632170 ENN71016.1 ERL89514.1 GEZM01001507 JAV97722.1 GDKW01001085 JAI55510.1 GBYB01011554 JAG81321.1 NEVH01014379 PNF27658.1 GBBI01002823 JAC15889.1 KQ981855 KYN34766.1 PYGN01000142 PSN53018.1 KQ977726 KYN00279.1 KQ976522 KYM82072.1 GL888217 EGI64727.1 GL447768 EFN85952.1 GEBQ01023662 JAT16315.1 GECL01000128 JAP05996.1 GECZ01000659 JAS69110.1 KQ762400 OAD55907.1 GECU01010460 JAS97246.1 GEDC01011818 GEDC01002843 JAS25480.1 JAS34455.1 GBGD01003494 JAC85395.1 KQ983219 KYQ46525.1 KQ980989 KYN10551.1 KQ971331 EFA01007.1 KQ435987 KOX67731.1 LJIG01016121 KRT81553.1 GBHO01040151 GBRD01004793 JAG03453.1 JAG61028.1 ACPB03021089 GL435626 EFN72984.1 KK852714 KDR17916.1 GACK01006500 JAA58534.1 GEDV01006407 JAP82150.1 GEFH01004836 JAP63745.1 GGLE01004189 MBY08315.1 LBMM01012351 KMQ86269.1 GL732525 EFX89016.1 GBZX01002638 JAG90102.1 GFPF01008525 MAA19671.1 GDIQ01167791 JAK83934.1 GBBL01002315 JAC25005.1 NEDP02001639 OWF52796.1 GFAC01006912 JAT92276.1 GFAA01000671 JAU02764.1 GBBK01004301 JAC20181.1 KK107020 QOIP01000006 EZA62601.1 RLU22014.1 ADTU01001292 LRGB01000024 KZS21682.1 GDIP01065766 JAM37949.1 GDIQ01028571 JAN66166.1 GADI01002977 JAA70831.1 ABJB010041971 DS661451 EEC03095.1 CP026255 AWP11955.1 GBYX01181532 JAO85724.1 HADW01004778 HADX01006462 SBP06178.1 BT082320 ACQ58027.1 HAEH01002431 HAEI01005152 SBR92007.1 HAED01008624 HAEE01007312 SBQ94836.1 HAEB01008107 HAEC01008748 SBQ76886.1 NHOQ01001766 PWA22238.1 HADZ01022201 HAEA01007924 SBQ36404.1 HADY01005626 HAEJ01007289 SBP44111.1 GG666648 EEN46146.1 GDIP01243510 JAI79891.1 AFYH01015977
JAT89456.1 GAIX01013377 JAA79183.1 KQ461190 KQ460205 KPJ07138.1 KPJ16799.1 KQ459590 KPI97408.1 AGBW02008299 OWR53791.1 ODYU01006722 SOQ48825.1 KQ414578 KOC71189.1 KZ288253 PBC31001.1 APGK01057714 KB741280 KB632170 ENN71016.1 ERL89514.1 GEZM01001507 JAV97722.1 GDKW01001085 JAI55510.1 GBYB01011554 JAG81321.1 NEVH01014379 PNF27658.1 GBBI01002823 JAC15889.1 KQ981855 KYN34766.1 PYGN01000142 PSN53018.1 KQ977726 KYN00279.1 KQ976522 KYM82072.1 GL888217 EGI64727.1 GL447768 EFN85952.1 GEBQ01023662 JAT16315.1 GECL01000128 JAP05996.1 GECZ01000659 JAS69110.1 KQ762400 OAD55907.1 GECU01010460 JAS97246.1 GEDC01011818 GEDC01002843 JAS25480.1 JAS34455.1 GBGD01003494 JAC85395.1 KQ983219 KYQ46525.1 KQ980989 KYN10551.1 KQ971331 EFA01007.1 KQ435987 KOX67731.1 LJIG01016121 KRT81553.1 GBHO01040151 GBRD01004793 JAG03453.1 JAG61028.1 ACPB03021089 GL435626 EFN72984.1 KK852714 KDR17916.1 GACK01006500 JAA58534.1 GEDV01006407 JAP82150.1 GEFH01004836 JAP63745.1 GGLE01004189 MBY08315.1 LBMM01012351 KMQ86269.1 GL732525 EFX89016.1 GBZX01002638 JAG90102.1 GFPF01008525 MAA19671.1 GDIQ01167791 JAK83934.1 GBBL01002315 JAC25005.1 NEDP02001639 OWF52796.1 GFAC01006912 JAT92276.1 GFAA01000671 JAU02764.1 GBBK01004301 JAC20181.1 KK107020 QOIP01000006 EZA62601.1 RLU22014.1 ADTU01001292 LRGB01000024 KZS21682.1 GDIP01065766 JAM37949.1 GDIQ01028571 JAN66166.1 GADI01002977 JAA70831.1 ABJB010041971 DS661451 EEC03095.1 CP026255 AWP11955.1 GBYX01181532 JAO85724.1 HADW01004778 HADX01006462 SBP06178.1 BT082320 ACQ58027.1 HAEH01002431 HAEI01005152 SBR92007.1 HAED01008624 HAEE01007312 SBQ94836.1 HAEB01008107 HAEC01008748 SBQ76886.1 NHOQ01001766 PWA22238.1 HADZ01022201 HAEA01007924 SBQ36404.1 HADY01005626 HAEJ01007289 SBP44111.1 GG666648 EEN46146.1 GDIP01243510 JAI79891.1 AFYH01015977
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000192223 UP000053825 UP000242457 UP000005203 UP000019118 UP000030742 UP000235965 UP000078541 UP000245037 UP000078542 UP000078540 UP000007755 UP000008237 UP000075809 UP000078492 UP000007266 UP000053105 UP000002358 UP000015103 UP000000311 UP000027135 UP000036403 UP000000305 UP000242188 UP000085678 UP000053097 UP000279307 UP000005205 UP000261640 UP000076858 UP000264800 UP000007635 UP000265040 UP000261520 UP000257160 UP000265080 UP000261580 UP000257200 UP000264820 UP000265020 UP000001555 UP000246464 UP000192220 UP000264840 UP000265100 UP000265160 UP000245300 UP000261480 UP000242638 UP000001554 UP000002852 UP000261380 UP000265120 UP000008672
UP000192223 UP000053825 UP000242457 UP000005203 UP000019118 UP000030742 UP000235965 UP000078541 UP000245037 UP000078542 UP000078540 UP000007755 UP000008237 UP000075809 UP000078492 UP000007266 UP000053105 UP000002358 UP000015103 UP000000311 UP000027135 UP000036403 UP000000305 UP000242188 UP000085678 UP000053097 UP000279307 UP000005205 UP000261640 UP000076858 UP000264800 UP000007635 UP000265040 UP000261520 UP000257160 UP000265080 UP000261580 UP000257200 UP000264820 UP000265020 UP000001555 UP000246464 UP000192220 UP000264840 UP000265100 UP000265160 UP000245300 UP000261480 UP000242638 UP000001554 UP000002852 UP000261380 UP000265120 UP000008672
PRIDE
Pfam
PF03966 Trm112p
ProteinModelPortal
H9JQG5
A0A0L7LCR1
A0A2A4K5S7
A0A1E1WRK1
S4NUB3
A0A194RH80
+ More
A0A194PWM1 A0A212FJ82 A0A2H1W6U8 A0A1W4XGW0 A0A0L7RK31 A0A2A3EJ01 A0A088AQL9 N6TRU2 A0A1Y1NII6 A0A0P4W1G5 A0A0C9R5G8 A0A2J7QGF1 A0A023F4E5 A0A195F305 A0A2P8Z947 A0A151IG83 A0A195BCY4 F4WM83 E2BEA7 A0A1B6KY32 A0A0V0GDK3 A0A1B6H3A3 A0A310SNI7 A0A1B6JDB0 A0A1B6DIN9 A0A069DNL4 A0A151WFG5 A0A195DCD7 D6WHM6 A0A0N0BBK7 A0A0T6B2T4 A0A0A9WAC5 K7J0T0 T1I9K8 E2A0T1 A0A067R6E6 L7M605 A0A131YUI8 A0A131XCA8 A0A2R5LFV5 A0A0J7K7N4 E9FVZ3 A0A0C9SAE9 A0A224YYI2 A0A0P5LPI4 A0A023FWG6 A0A210QVU1 A0A1E1WZQ7 A0A1E1XU69 A0A023FDY7 A0A1S3HUW0 A0A026X3Q0 A0A158P2V8 A0A3Q3SWX1 A0A162SW91 A0A3Q2ZMR2 G3PXS1 A0A0P5XY68 A0A3Q1ITL6 A0A0P6GUF2 A0A3B3ZNH5 A0A3Q1BE39 A0A3P8SM81 A0A3Q4GDN3 A0A0K8RIC8 G3PXS4 A0A3Q1HS00 A0A3Q2Z8W9 A0A3Q2DU95 B7P923 A0A2U9C5T1 A0A0S7L872 A0A1A7WK78 A0A2I4B0M8 A0A3Q2VIW6 A0A3P8Q9J9 A0A3P9B990 A0A2I4B0P6 S4RJ54 C3KHA5 A0A1A8QF48 A0A1A8IBW6 A0A1A8GYF9 A0A3B3Y8G5 A0A3P9PL45 A0A315VFM9 A0A1A8DU99 A0A1A7ZP72 C3ZN06 A0A3B5QA98 A0A3B5KTH5 A0A0P4XD63 A0A3P8V317 H3BEP6
A0A194PWM1 A0A212FJ82 A0A2H1W6U8 A0A1W4XGW0 A0A0L7RK31 A0A2A3EJ01 A0A088AQL9 N6TRU2 A0A1Y1NII6 A0A0P4W1G5 A0A0C9R5G8 A0A2J7QGF1 A0A023F4E5 A0A195F305 A0A2P8Z947 A0A151IG83 A0A195BCY4 F4WM83 E2BEA7 A0A1B6KY32 A0A0V0GDK3 A0A1B6H3A3 A0A310SNI7 A0A1B6JDB0 A0A1B6DIN9 A0A069DNL4 A0A151WFG5 A0A195DCD7 D6WHM6 A0A0N0BBK7 A0A0T6B2T4 A0A0A9WAC5 K7J0T0 T1I9K8 E2A0T1 A0A067R6E6 L7M605 A0A131YUI8 A0A131XCA8 A0A2R5LFV5 A0A0J7K7N4 E9FVZ3 A0A0C9SAE9 A0A224YYI2 A0A0P5LPI4 A0A023FWG6 A0A210QVU1 A0A1E1WZQ7 A0A1E1XU69 A0A023FDY7 A0A1S3HUW0 A0A026X3Q0 A0A158P2V8 A0A3Q3SWX1 A0A162SW91 A0A3Q2ZMR2 G3PXS1 A0A0P5XY68 A0A3Q1ITL6 A0A0P6GUF2 A0A3B3ZNH5 A0A3Q1BE39 A0A3P8SM81 A0A3Q4GDN3 A0A0K8RIC8 G3PXS4 A0A3Q1HS00 A0A3Q2Z8W9 A0A3Q2DU95 B7P923 A0A2U9C5T1 A0A0S7L872 A0A1A7WK78 A0A2I4B0M8 A0A3Q2VIW6 A0A3P8Q9J9 A0A3P9B990 A0A2I4B0P6 S4RJ54 C3KHA5 A0A1A8QF48 A0A1A8IBW6 A0A1A8GYF9 A0A3B3Y8G5 A0A3P9PL45 A0A315VFM9 A0A1A8DU99 A0A1A7ZP72 C3ZN06 A0A3B5QA98 A0A3B5KTH5 A0A0P4XD63 A0A3P8V317 H3BEP6
PDB
6K0X
E-value=1.74765e-26,
Score=289
Ontologies
PANTHER
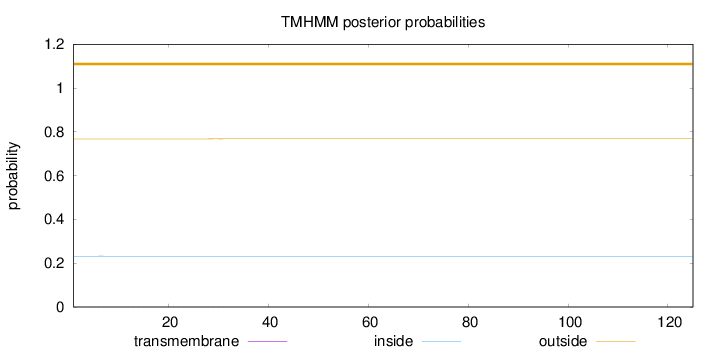
Topology
Length:
125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01372
Exp number, first 60 AAs:
0.01372
Total prob of N-in:
0.23217
outside
1 - 125
Population Genetic Test Statistics
Pi
53.625751
Theta
137.58373
Tajima's D
-1.964707
CLR
1161.637589
CSRT
0.0170991450427479
Interpretation
Uncertain
