Gene
KWMTBOMO06657 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011774
Annotation
PREDICTED:_purine_nucleoside_phosphorylase-like_isoform_X2_[Amyelois_transitella]
Full name
Purine nucleoside phosphorylase
Alternative Name
Inosine-guanosine phosphorylase
Location in the cell
Cytoplasmic Reliability : 3.355
Sequence
CDS
ATGGCACCTATAAACGCGAACGATATCGTGGAGAAAAATTGTTTGGTCACGCAGAAGGTCCTGCCGGAAATCGGATCTGATTGTAATGGAAATGAAAAAACTGGATATTCTTATGAAACATTAGTGGAGACCGCAAATTTCTTGCTGTCGAGAATATCAGAGAAACCGAACATTGGGATCATTTGCGGCTCTGGGATGGGTTCACTAGCAGAAAGTATAACAGACGGCGTAAGAATACCATACGAAGATATTCCAAACTTTCCCATAAGCACAGTGGAGGGTCATCATGGTCAGCTTGTTTTCGGTCACATAGAAGGAGTGTCCGTCGTTGCTATGCAGGGCCGTTTTCATTACTACGAAGGATATCCACTGTGGAAGTGTTGTCTACCAGTGCGAGTGATGAAACTGTTGGGAGTGAAAATTTTAATAGCGACTAACGCTGCAGGCGGCCTTAACCCTAACTACAAAATTGGTGATCTGATGATTGTAAGAGACCACATCAACATGATGGGTTTTGCTGGAAACAATCCTTTGCATGGACCCAATGACGAGAGGTTTGGACCTAGATTCCCCCCTATGAACAAAGCCTATAATTATGAATTTAGAAAAATTGCTAAAGAGGTTGCGAAGGAATTAAACATCGATCATATTGTAAGGGAAGGAGTTTATACATGTTTGGGCGGTCCAAATTTCGAGACAGTCGCCGAATTGAATATGTTAAAGATGGTTGGAGTAGACGCTGTTGGCATGTCTACTGTTCACGAGGTTATCACAGCTAGACACTGCGACATTAAAGTATTCGGATTATCTCTGATTACGAATGAATGCATTACTAATTACGATCAAGATGCAGAAGCTAATCACGAGGAAGTTCTTGACGTAGGCCGTATGAGGCAGGAAATGCTCCGAAAATATGTATCCTGTCTTGTTAAGAAATTCTCACAGGCCGACCCTGATGCACCATGA
Protein
MAPINANDIVEKNCLVTQKVLPEIGSDCNGNEKTGYSYETLVETANFLLSRISEKPNIGIICGSGMGSLAESITDGVRIPYEDIPNFPISTVEGHHGQLVFGHIEGVSVVAMQGRFHYYEGYPLWKCCLPVRVMKLLGVKILIATNAAGGLNPNYKIGDLMIVRDHINMMGFAGNNPLHGPNDERFGPRFPPMNKAYNYEFRKIAKEVAKELNIDHIVREGVYTCLGGPNFETVAELNMLKMVGVDAVGMSTVHEVITARHCDIKVFGLSLITNECITNYDQDAEANHEEVLDVGRMRQEMLRKYVSCLVKKFSQADPDAP
Summary
Description
The purine nucleoside phosphorylases catalyze the phosphorolytic breakdown of the N-glycosidic bond in the beta-(deoxy)ribonucleoside molecules, with the formation of the corresponding free purine bases and pentose-1-phosphate.
Catalytic Activity
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha-D-ribose 1-phosphate
Similarity
Belongs to the PNP/MTAP phosphorylase family.
Uniprot
H9JQG8
A0A2A4K6V6
A0A3S2L924
A0A194QUB6
I4DJX3
A0A212FG34
+ More
A0A2H1WEJ7 A0A194PXB1 I4DN26 F4X5C6 A0A151IRC0 A0A151WRP6 A0A151JXW1 A0A026WGL7 A0A336MF31 A0A2A3EEE2 V9ID02 V5G3C0 D6WHJ8 E2A5F2 A0A0C9QZ53 A0A1Y1MAZ9 V9IB69 V5I8X0 A0A088ARK6 A0A158NMI3 A0A195CVN9 A0A195AZQ9 A0A1Y1MDD8 A0A0M9A8P0 A0A1I8MDJ9 A0A0L0CHI9 A0A1I8MDK1 A0A1I8MDK5 T1PEF0 A0A1L8EFX7 A0A0A1WFJ0 W8C2C0 A0A1L8EFV6 A0A1L8E9B1 W8BG08 A0A0A1XDW8 A0A1I8PPA7 A0A3Q0IZV9 A0A2J7QGQ8 A0A1B6FLG7 A0A1A9XPE4 A0A0K8W5M4 K7IM21 A0A1A9VN56 A0A1A9WC80 D3TLL2 A0A034VEC1 A0A0K8V1L9 A0A1J1IK46 A0A034VGE2 A0A1B0AQ94 A0A232EJE3 A0A0K8V0D3 A0A2J7QGQ4 A0A1A9ZML4 A0A0K8V931 A0A023GHV8 A0A154P2N4 A0A1B6GSG4 N6U0W3 T1GD09 A0A1E1XFC6 A0A224Z2T0 G3MGH6 W5JNR5 A0A131YZ73 A0A067QW09 A0A224YXB2 A0A023FYA5 A0A131YDI9 T1J2B2 A0A2M4CME6 A0A2M4CMC9 A0A131XMM8 A0A0L7RIS9 A0A023FJU0 A0A2M4A309 A0A2M4A2J4 A0A3R7M3L2 U5EV37 A0A2M4BSI9 A0A0P4W8N2 A0A0N7ZCN7 A0A0P4WGT2 A0A2M3Z5F4 A0A2M4BW19 A0A084WA99 A0A0P5KMY5 A0A1Q3FGB9 A0A1Q3FGE3 A0A0N8ATM5 A0A0P5RJ47 A0A0P5JKK0 E9GU60
A0A2H1WEJ7 A0A194PXB1 I4DN26 F4X5C6 A0A151IRC0 A0A151WRP6 A0A151JXW1 A0A026WGL7 A0A336MF31 A0A2A3EEE2 V9ID02 V5G3C0 D6WHJ8 E2A5F2 A0A0C9QZ53 A0A1Y1MAZ9 V9IB69 V5I8X0 A0A088ARK6 A0A158NMI3 A0A195CVN9 A0A195AZQ9 A0A1Y1MDD8 A0A0M9A8P0 A0A1I8MDJ9 A0A0L0CHI9 A0A1I8MDK1 A0A1I8MDK5 T1PEF0 A0A1L8EFX7 A0A0A1WFJ0 W8C2C0 A0A1L8EFV6 A0A1L8E9B1 W8BG08 A0A0A1XDW8 A0A1I8PPA7 A0A3Q0IZV9 A0A2J7QGQ8 A0A1B6FLG7 A0A1A9XPE4 A0A0K8W5M4 K7IM21 A0A1A9VN56 A0A1A9WC80 D3TLL2 A0A034VEC1 A0A0K8V1L9 A0A1J1IK46 A0A034VGE2 A0A1B0AQ94 A0A232EJE3 A0A0K8V0D3 A0A2J7QGQ4 A0A1A9ZML4 A0A0K8V931 A0A023GHV8 A0A154P2N4 A0A1B6GSG4 N6U0W3 T1GD09 A0A1E1XFC6 A0A224Z2T0 G3MGH6 W5JNR5 A0A131YZ73 A0A067QW09 A0A224YXB2 A0A023FYA5 A0A131YDI9 T1J2B2 A0A2M4CME6 A0A2M4CMC9 A0A131XMM8 A0A0L7RIS9 A0A023FJU0 A0A2M4A309 A0A2M4A2J4 A0A3R7M3L2 U5EV37 A0A2M4BSI9 A0A0P4W8N2 A0A0N7ZCN7 A0A0P4WGT2 A0A2M3Z5F4 A0A2M4BW19 A0A084WA99 A0A0P5KMY5 A0A1Q3FGB9 A0A1Q3FGE3 A0A0N8ATM5 A0A0P5RJ47 A0A0P5JKK0 E9GU60
EC Number
2.4.2.1
Pubmed
EMBL
BABH01011972
BABH01011973
BABH01011974
NWSH01000114
PCG79402.1
RSAL01000448
+ More
RVE41679.1 KQ461190 KPJ07146.1 AK401591 BAM18213.1 AGBW02008724 OWR52690.1 ODYU01008119 SOQ51447.1 KQ459590 KPI97399.1 AK402694 BAM19316.1 GL888711 EGI58340.1 KQ981130 KYN09268.1 KQ982797 KYQ50582.1 KQ981550 KYN40023.1 KK107238 QOIP01000007 EZA54836.1 RLU20810.1 UFQT01001029 UFQT01002424 SSX28540.1 SSX33425.1 KZ288282 PBC29589.1 JR038612 AEY58341.1 GALX01003951 JAB64515.1 KQ971331 EFA01001.1 GL436926 EFN71333.1 GBYB01001005 JAG70772.1 GEZM01036044 JAV83009.1 JR038611 AEY58340.1 GALX01003950 JAB64516.1 ADTU01020586 ADTU01020587 ADTU01020588 KQ977231 KYN04741.1 KQ976697 KYM77520.1 GEZM01036045 JAV83008.1 KQ435726 KOX78203.1 JRES01000390 KNC31720.1 KA646495 AFP61124.1 GFDG01001274 JAV17525.1 GBXI01017114 GBXI01010393 JAC97177.1 JAD03899.1 GAMC01010671 JAB95884.1 GFDG01001273 JAV17526.1 GFDG01003613 JAV15186.1 GAMC01010672 JAB95883.1 GBXI01005634 JAD08658.1 NEVH01014359 PNF27771.1 GECZ01018750 JAS51019.1 GDHF01005893 GDHF01002942 JAI46421.1 JAI49372.1 CCAG010019517 CCAG010019518 EZ422314 ADD18590.1 GAKP01018108 JAC40844.1 GDHF01019510 JAI32804.1 CVRI01000050 CRK99446.1 GAKP01018107 JAC40845.1 JXJN01001758 NNAY01004028 OXU18474.1 GDHF01020076 JAI32238.1 PNF27772.1 GDHF01016932 JAI35382.1 GBBM01001916 JAC33502.1 KQ434806 KZC06117.1 GECZ01004399 JAS65370.1 APGK01053843 KB741237 ENN72187.1 CAQQ02024629 CAQQ02024630 CAQQ02024631 CAQQ02024632 GFAC01001228 JAT97960.1 GFPF01011123 MAA22269.1 JO840977 AEO32594.1 ADMH02000830 ETN64943.1 GEDV01004705 JAP83852.1 KK852879 KDR14468.1 GFPF01011122 MAA22268.1 GBBL01001592 JAC25728.1 GEDV01011939 JAP76618.1 JH431796 GGFL01002334 MBW66512.1 GGFL01002265 MBW66443.1 GEFH01000178 JAP68403.1 KQ414584 KOC70646.1 GBBK01003167 JAC21315.1 GGFK01001707 MBW35028.1 GGFK01001641 MBW34962.1 QCYY01002961 ROT66297.1 GANO01002043 JAB57828.1 GGFJ01006904 MBW56045.1 GDRN01063561 JAI64958.1 GDRN01063559 JAI64960.1 GDRN01063560 JAI64959.1 GGFM01002984 MBW23735.1 GGFJ01008118 MBW57259.1 ATLV01022056 KE525327 KFB47143.1 GDIQ01189274 JAK62451.1 GFDL01008448 JAV26597.1 GFDL01008430 JAV26615.1 GDIQ01244059 JAK07666.1 GDIQ01186321 GDIQ01119202 GDIP01116511 LRGB01002384 JAL32524.1 JAL87203.1 KZS07994.1 GDIQ01201776 GDIQ01189275 JAK49949.1 GL732565 EFX76980.1
RVE41679.1 KQ461190 KPJ07146.1 AK401591 BAM18213.1 AGBW02008724 OWR52690.1 ODYU01008119 SOQ51447.1 KQ459590 KPI97399.1 AK402694 BAM19316.1 GL888711 EGI58340.1 KQ981130 KYN09268.1 KQ982797 KYQ50582.1 KQ981550 KYN40023.1 KK107238 QOIP01000007 EZA54836.1 RLU20810.1 UFQT01001029 UFQT01002424 SSX28540.1 SSX33425.1 KZ288282 PBC29589.1 JR038612 AEY58341.1 GALX01003951 JAB64515.1 KQ971331 EFA01001.1 GL436926 EFN71333.1 GBYB01001005 JAG70772.1 GEZM01036044 JAV83009.1 JR038611 AEY58340.1 GALX01003950 JAB64516.1 ADTU01020586 ADTU01020587 ADTU01020588 KQ977231 KYN04741.1 KQ976697 KYM77520.1 GEZM01036045 JAV83008.1 KQ435726 KOX78203.1 JRES01000390 KNC31720.1 KA646495 AFP61124.1 GFDG01001274 JAV17525.1 GBXI01017114 GBXI01010393 JAC97177.1 JAD03899.1 GAMC01010671 JAB95884.1 GFDG01001273 JAV17526.1 GFDG01003613 JAV15186.1 GAMC01010672 JAB95883.1 GBXI01005634 JAD08658.1 NEVH01014359 PNF27771.1 GECZ01018750 JAS51019.1 GDHF01005893 GDHF01002942 JAI46421.1 JAI49372.1 CCAG010019517 CCAG010019518 EZ422314 ADD18590.1 GAKP01018108 JAC40844.1 GDHF01019510 JAI32804.1 CVRI01000050 CRK99446.1 GAKP01018107 JAC40845.1 JXJN01001758 NNAY01004028 OXU18474.1 GDHF01020076 JAI32238.1 PNF27772.1 GDHF01016932 JAI35382.1 GBBM01001916 JAC33502.1 KQ434806 KZC06117.1 GECZ01004399 JAS65370.1 APGK01053843 KB741237 ENN72187.1 CAQQ02024629 CAQQ02024630 CAQQ02024631 CAQQ02024632 GFAC01001228 JAT97960.1 GFPF01011123 MAA22269.1 JO840977 AEO32594.1 ADMH02000830 ETN64943.1 GEDV01004705 JAP83852.1 KK852879 KDR14468.1 GFPF01011122 MAA22268.1 GBBL01001592 JAC25728.1 GEDV01011939 JAP76618.1 JH431796 GGFL01002334 MBW66512.1 GGFL01002265 MBW66443.1 GEFH01000178 JAP68403.1 KQ414584 KOC70646.1 GBBK01003167 JAC21315.1 GGFK01001707 MBW35028.1 GGFK01001641 MBW34962.1 QCYY01002961 ROT66297.1 GANO01002043 JAB57828.1 GGFJ01006904 MBW56045.1 GDRN01063561 JAI64958.1 GDRN01063559 JAI64960.1 GDRN01063560 JAI64959.1 GGFM01002984 MBW23735.1 GGFJ01008118 MBW57259.1 ATLV01022056 KE525327 KFB47143.1 GDIQ01189274 JAK62451.1 GFDL01008448 JAV26597.1 GFDL01008430 JAV26615.1 GDIQ01244059 JAK07666.1 GDIQ01186321 GDIQ01119202 GDIP01116511 LRGB01002384 JAL32524.1 JAL87203.1 KZS07994.1 GDIQ01201776 GDIQ01189275 JAK49949.1 GL732565 EFX76980.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000007755 UP000078492 UP000075809 UP000078541 UP000053097 UP000279307 UP000242457 UP000007266 UP000000311 UP000005203 UP000005205 UP000078542 UP000078540 UP000053105 UP000095301 UP000037069 UP000095300 UP000079169 UP000235965 UP000092443 UP000002358 UP000078200 UP000091820 UP000092444 UP000183832 UP000092460 UP000215335 UP000092445 UP000076502 UP000019118 UP000015102 UP000000673 UP000027135 UP000053825 UP000283509 UP000030765 UP000076858 UP000000305
UP000007755 UP000078492 UP000075809 UP000078541 UP000053097 UP000279307 UP000242457 UP000007266 UP000000311 UP000005203 UP000005205 UP000078542 UP000078540 UP000053105 UP000095301 UP000037069 UP000095300 UP000079169 UP000235965 UP000092443 UP000002358 UP000078200 UP000091820 UP000092444 UP000183832 UP000092460 UP000215335 UP000092445 UP000076502 UP000019118 UP000015102 UP000000673 UP000027135 UP000053825 UP000283509 UP000030765 UP000076858 UP000000305
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JQG8
A0A2A4K6V6
A0A3S2L924
A0A194QUB6
I4DJX3
A0A212FG34
+ More
A0A2H1WEJ7 A0A194PXB1 I4DN26 F4X5C6 A0A151IRC0 A0A151WRP6 A0A151JXW1 A0A026WGL7 A0A336MF31 A0A2A3EEE2 V9ID02 V5G3C0 D6WHJ8 E2A5F2 A0A0C9QZ53 A0A1Y1MAZ9 V9IB69 V5I8X0 A0A088ARK6 A0A158NMI3 A0A195CVN9 A0A195AZQ9 A0A1Y1MDD8 A0A0M9A8P0 A0A1I8MDJ9 A0A0L0CHI9 A0A1I8MDK1 A0A1I8MDK5 T1PEF0 A0A1L8EFX7 A0A0A1WFJ0 W8C2C0 A0A1L8EFV6 A0A1L8E9B1 W8BG08 A0A0A1XDW8 A0A1I8PPA7 A0A3Q0IZV9 A0A2J7QGQ8 A0A1B6FLG7 A0A1A9XPE4 A0A0K8W5M4 K7IM21 A0A1A9VN56 A0A1A9WC80 D3TLL2 A0A034VEC1 A0A0K8V1L9 A0A1J1IK46 A0A034VGE2 A0A1B0AQ94 A0A232EJE3 A0A0K8V0D3 A0A2J7QGQ4 A0A1A9ZML4 A0A0K8V931 A0A023GHV8 A0A154P2N4 A0A1B6GSG4 N6U0W3 T1GD09 A0A1E1XFC6 A0A224Z2T0 G3MGH6 W5JNR5 A0A131YZ73 A0A067QW09 A0A224YXB2 A0A023FYA5 A0A131YDI9 T1J2B2 A0A2M4CME6 A0A2M4CMC9 A0A131XMM8 A0A0L7RIS9 A0A023FJU0 A0A2M4A309 A0A2M4A2J4 A0A3R7M3L2 U5EV37 A0A2M4BSI9 A0A0P4W8N2 A0A0N7ZCN7 A0A0P4WGT2 A0A2M3Z5F4 A0A2M4BW19 A0A084WA99 A0A0P5KMY5 A0A1Q3FGB9 A0A1Q3FGE3 A0A0N8ATM5 A0A0P5RJ47 A0A0P5JKK0 E9GU60
A0A2H1WEJ7 A0A194PXB1 I4DN26 F4X5C6 A0A151IRC0 A0A151WRP6 A0A151JXW1 A0A026WGL7 A0A336MF31 A0A2A3EEE2 V9ID02 V5G3C0 D6WHJ8 E2A5F2 A0A0C9QZ53 A0A1Y1MAZ9 V9IB69 V5I8X0 A0A088ARK6 A0A158NMI3 A0A195CVN9 A0A195AZQ9 A0A1Y1MDD8 A0A0M9A8P0 A0A1I8MDJ9 A0A0L0CHI9 A0A1I8MDK1 A0A1I8MDK5 T1PEF0 A0A1L8EFX7 A0A0A1WFJ0 W8C2C0 A0A1L8EFV6 A0A1L8E9B1 W8BG08 A0A0A1XDW8 A0A1I8PPA7 A0A3Q0IZV9 A0A2J7QGQ8 A0A1B6FLG7 A0A1A9XPE4 A0A0K8W5M4 K7IM21 A0A1A9VN56 A0A1A9WC80 D3TLL2 A0A034VEC1 A0A0K8V1L9 A0A1J1IK46 A0A034VGE2 A0A1B0AQ94 A0A232EJE3 A0A0K8V0D3 A0A2J7QGQ4 A0A1A9ZML4 A0A0K8V931 A0A023GHV8 A0A154P2N4 A0A1B6GSG4 N6U0W3 T1GD09 A0A1E1XFC6 A0A224Z2T0 G3MGH6 W5JNR5 A0A131YZ73 A0A067QW09 A0A224YXB2 A0A023FYA5 A0A131YDI9 T1J2B2 A0A2M4CME6 A0A2M4CMC9 A0A131XMM8 A0A0L7RIS9 A0A023FJU0 A0A2M4A309 A0A2M4A2J4 A0A3R7M3L2 U5EV37 A0A2M4BSI9 A0A0P4W8N2 A0A0N7ZCN7 A0A0P4WGT2 A0A2M3Z5F4 A0A2M4BW19 A0A084WA99 A0A0P5KMY5 A0A1Q3FGB9 A0A1Q3FGE3 A0A0N8ATM5 A0A0P5RJ47 A0A0P5JKK0 E9GU60
PDB
2P4S
E-value=7.95132e-101,
Score=936
Ontologies
PATHWAY
GO
PANTHER
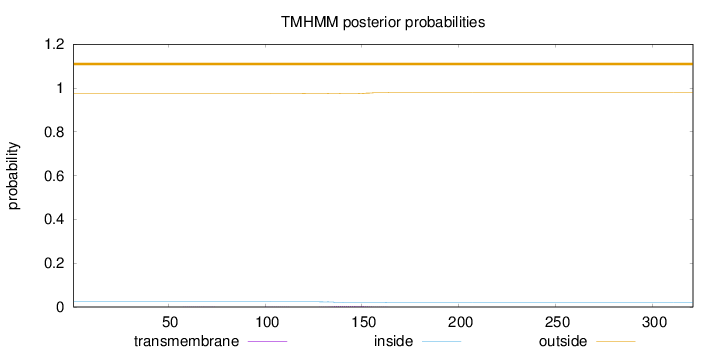
Topology
Length:
321
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09569
Exp number, first 60 AAs:
0.00064
Total prob of N-in:
0.02429
outside
1 - 321
Population Genetic Test Statistics
Pi
283.400321
Theta
190.576673
Tajima's D
1.663534
CLR
0.304513
CSRT
0.826308684565772
Interpretation
Uncertain
