Gene
KWMTBOMO06655
Pre Gene Modal
BGIBMGA011775
Annotation
PREDICTED:_tRNA-splicing_endonuclease_subunit_Sen34_[Papilio_xuthus]
Full name
tRNA-splicing endonuclease subunit Sen34
Location in the cell
Cytoplasmic Reliability : 2.107 Extracellular Reliability : 1.467
Sequence
CDS
ATGATTTCTTGGATCTTTGCAGGGCTTCCGATTGCATTATCTTCTGAAGAAACAGCATTATTGGTTGAAAAGAGGATATGTGAACTTCATGAAATGCCAGCTGAATTTTGTAAATATGAACCAACAGAAAAGGATAAAGAATTAATGGAAGAATTTTTAAAAAAAGTACTTGAACAACAAGCATCAGCATTAAAGAAACGCAAAATAGAACAATTATCACAAAAGATTGATATCATTGTGGCTGGGAAAAAGAAAAACCTCATAAATAAAGGGATGTCTGATGCAAATATTGATAAAGATGCAATGTTAGAAGAAGAAATAAAAAGAATACAAGATTTAGATCCAGACCACACTTTAGTACAACTTCCACAAGAAATGTATAGAAATATAGAAACCAGTCCAGTAGGTTTGGATGTATTAAGACCAAATATTTTGGAAGGAGATGGTGCCGTTAAGTATTCTATATTCAAAGACTTATGGGAGAAAGGATATTATGTTACCAGTGGATCAAAATTTGGATGTGATTATCTTATATACCCAGGCGATCCTGTGCAGTTTCATGCCTCGCATATGGTTCGATGCGTTTGTAATCAAGAAAATGTTTTCCACCCTAAAAGCTTAGTGGCGTTCGGCAGATTATCAGTGGCCGTCAATAAGTATGCAGTTTTTGCATATAGAACTCAGTGTGCGCGAATTGCATATCAAACTCTGCAATGGCATGACAGCATGGATTCTTGA
Protein
MISWIFAGLPIALSSEETALLVEKRICELHEMPAEFCKYEPTEKDKELMEEFLKKVLEQQASALKKRKIEQLSQKIDIIVAGKKKNLINKGMSDANIDKDAMLEEEIKRIQDLDPDHTLVQLPQEMYRNIETSPVGLDVLRPNILEGDGAVKYSIFKDLWEKGYYVTSGSKFGCDYLIYPGDPVQFHASHMVRCVCNQENVFHPKSLVAFGRLSVAVNKYAVFAYRTQCARIAYQTLQWHDSMDS
Summary
Description
Constitutes one of the two catalytic subunit of the tRNA-splicing endonuclease complex, a complex responsible for identification and cleavage of the splice sites in pre-tRNA. It cleaves pre-tRNA at the 5'- and 3'-splice sites to release the intron. The products are an intron and two tRNA half-molecules bearing 2',3'-cyclic phosphate and 5'-OH termini. There are no conserved sequences at the splice sites, but the intron is invariably located at the same site in the gene, placing the splice sites an invariant distance from the constant structural features of the tRNA body.
Catalytic Activity
PretRNA = a 3'-half-tRNA molecule with a 5'-OH end + a 5'-half-tRNA molecule with a 2',3'-cyclic phosphate end + an intron with a 2',3'-cyclic phosphate and a 5'-hydroxyl terminus.
Similarity
Belongs to the tRNA-intron endonuclease family.
Uniprot
H9JQG9
A0A2H1WEF9
A0A2A4K666
A0A194PVP3
A0A1E1W6P0
A0A194QNP7
+ More
S4PDN4 A0A3S2N524 A0A0L7LV47 A0A067QI60 A0A1B0DJ44 B0X2T9 B4QK26 A0A1W4XHE4 B4HHS9 A0A2J7RA58 A0A2J7RA46 A0A1B0CSR0 A0A026W1B4 A0A1Q3F5L3 A0A1A9VFD4 Q0E8E7 D2NUI2 G4LU02 A0A1B0G9Y8 A0A1B0A2A3 A0A182GG98 A0A0M4EM16 E2AUD7 A0A182G1R5 A0A1S4FJZ8 A0A2A3E1E7 A0A088AVZ8 A0A1B6CWN3 A0A1I8P9N7 A0A1I8P9W0 A0A1B0AVW0 Q16YD2 B5DQS3 A0A154PDJ4 A0A182WDX8 B3MAW2 A0A1W4UKM1 B4PIG6 A0A1A9XR90 A0A1W4U8S9 E9J1N6 A0A182IQJ8 A0A232EU37 A0A0N8B842 A0A0P5Y840 A0A0P6FM94 A0A158P3X0 A0A1I8MDV3 A0A0Q9XG26 W8BUP8 A0A182TA15 B4KVK9 A0A195BKM6 D6WIR4 K7IQN6 A0A182Q2X7 B4J3T7 B4LF54 A0NGQ6 A0A034WRL9 A0A0K8W0B3 A0A151J8Z8 A0A182L517 E0VW20 A0A3B0J386 A0A195CNX8 A0A182HHI2 A0A182MHV5 A0A182UAP6 A0A151WVB4 A0A0L0CED2 A0A182VGL5 A0A182RRZ6 A0A084WIP1 F4WKV0 A0A182XTJ4 A0A182JRB0 A0A0P6HN96 E9FVZ2 A0A0P5LIF7 A0A182N2W8 B3ND56 A0A0P6AJ82 A0A1Y1LF38 A0A0A1WV44 A0A1B6M6M6 A0A0A9ZH52 A0A0K8SKM5 A0A2T7PFP2 A0A2M4AYD3 A0A2M4CUJ4 A0A2K6VAZ7 A0A0L7QPN5
S4PDN4 A0A3S2N524 A0A0L7LV47 A0A067QI60 A0A1B0DJ44 B0X2T9 B4QK26 A0A1W4XHE4 B4HHS9 A0A2J7RA58 A0A2J7RA46 A0A1B0CSR0 A0A026W1B4 A0A1Q3F5L3 A0A1A9VFD4 Q0E8E7 D2NUI2 G4LU02 A0A1B0G9Y8 A0A1B0A2A3 A0A182GG98 A0A0M4EM16 E2AUD7 A0A182G1R5 A0A1S4FJZ8 A0A2A3E1E7 A0A088AVZ8 A0A1B6CWN3 A0A1I8P9N7 A0A1I8P9W0 A0A1B0AVW0 Q16YD2 B5DQS3 A0A154PDJ4 A0A182WDX8 B3MAW2 A0A1W4UKM1 B4PIG6 A0A1A9XR90 A0A1W4U8S9 E9J1N6 A0A182IQJ8 A0A232EU37 A0A0N8B842 A0A0P5Y840 A0A0P6FM94 A0A158P3X0 A0A1I8MDV3 A0A0Q9XG26 W8BUP8 A0A182TA15 B4KVK9 A0A195BKM6 D6WIR4 K7IQN6 A0A182Q2X7 B4J3T7 B4LF54 A0NGQ6 A0A034WRL9 A0A0K8W0B3 A0A151J8Z8 A0A182L517 E0VW20 A0A3B0J386 A0A195CNX8 A0A182HHI2 A0A182MHV5 A0A182UAP6 A0A151WVB4 A0A0L0CED2 A0A182VGL5 A0A182RRZ6 A0A084WIP1 F4WKV0 A0A182XTJ4 A0A182JRB0 A0A0P6HN96 E9FVZ2 A0A0P5LIF7 A0A182N2W8 B3ND56 A0A0P6AJ82 A0A1Y1LF38 A0A0A1WV44 A0A1B6M6M6 A0A0A9ZH52 A0A0K8SKM5 A0A2T7PFP2 A0A2M4AYD3 A0A2M4CUJ4 A0A2K6VAZ7 A0A0L7QPN5
EC Number
4.6.1.16
Pubmed
19121390
26354079
23622113
26227816
24845553
17994087
+ More
22936249 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 20798317 17510324 15632085 17550304 21282665 28648823 21347285 25315136 24495485 18057021 18362917 19820115 20075255 12364791 14747013 17210077 25348373 20966253 20566863 26108605 24438588 21719571 21292972 28004739 25830018 25401762 26823975 20920257
22936249 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 20798317 17510324 15632085 17550304 21282665 28648823 21347285 25315136 24495485 18057021 18362917 19820115 20075255 12364791 14747013 17210077 25348373 20966253 20566863 26108605 24438588 21719571 21292972 28004739 25830018 25401762 26823975 20920257
EMBL
BABH01011976
ODYU01008119
SOQ51449.1
NWSH01000114
PCG79404.1
KQ459590
+ More
KPI97397.1 GDQN01008392 JAT82662.1 KQ461190 KPJ07148.1 GAIX01003601 JAA88959.1 RSAL01000448 RVE41681.1 JTDY01000028 KOB79332.1 KK853353 KDR08208.1 AJVK01006199 DS232298 EDS39413.1 CM000363 CM002912 EDX10464.1 KMY99621.1 CH480815 EDW41494.1 NEVH01006578 PNF37704.1 PNF37705.1 AJWK01026423 KK107570 QOIP01000005 EZA48854.1 RLU23094.1 GFDL01012199 JAV22846.1 AE014296 ABI31252.2 BT120125 ADB46406.1 BT132632 AEQ05528.1 CCAG010008280 JXUM01061327 KQ562145 KXJ76577.1 CP012525 ALC44610.1 GL442815 EFN62994.1 JXUM01137733 KQ568620 KXJ68944.1 KZ288451 PBC25573.1 GEDC01019705 JAS17593.1 JXJN01004400 CH477518 EAT39638.1 CH379069 EDY73389.2 KQ434878 KZC09901.1 CH902618 EDV39196.1 CM000159 EDW94523.1 GL767674 EFZ13246.1 NNAY01002176 OXU21868.1 GDIP01146506 GDIQ01203523 LRGB01000024 JAJ76896.1 JAK48202.1 KZS21683.1 GDIP01061564 JAM42151.1 GDIQ01048233 JAN46504.1 ADTU01008608 CH933809 KRG06649.1 GAMC01001475 JAC05081.1 EDW19480.1 KQ976453 KYM85324.1 KQ971321 EEZ99511.2 AXCN02001090 CH916366 EDV97318.1 CH940647 EDW70242.2 AAAB01008986 EAU75852.2 GAKP01001678 JAC57274.1 GDHF01007773 JAI44541.1 KQ979480 KYN21264.1 AAZO01005737 DS235816 EEB17576.1 OUUW01000002 SPP76054.1 KQ977513 KYN02182.1 APCN01002416 AXCM01009764 KQ982708 KYQ51803.1 JRES01000502 KNC30606.1 ATLV01023941 KE525347 KFB50085.1 GL888206 EGI65192.1 GDIQ01016728 JAN78009.1 GL732525 EFX88635.1 GDIQ01171333 JAK80392.1 CH954178 EDV51778.1 GDIP01030112 JAM73603.1 GEZM01062527 GEZM01062524 GEZM01062523 GEZM01062520 JAV69627.1 GBXI01012014 JAD02278.1 GEBQ01008409 JAT31568.1 GBHO01000591 JAG43013.1 GBRD01011958 GDHC01002291 JAG53866.1 JAQ16338.1 PZQS01000004 PVD32238.1 GGFK01012401 MBW45722.1 GGFL01004812 MBW68990.1 KQ414815 KOC60519.1
KPI97397.1 GDQN01008392 JAT82662.1 KQ461190 KPJ07148.1 GAIX01003601 JAA88959.1 RSAL01000448 RVE41681.1 JTDY01000028 KOB79332.1 KK853353 KDR08208.1 AJVK01006199 DS232298 EDS39413.1 CM000363 CM002912 EDX10464.1 KMY99621.1 CH480815 EDW41494.1 NEVH01006578 PNF37704.1 PNF37705.1 AJWK01026423 KK107570 QOIP01000005 EZA48854.1 RLU23094.1 GFDL01012199 JAV22846.1 AE014296 ABI31252.2 BT120125 ADB46406.1 BT132632 AEQ05528.1 CCAG010008280 JXUM01061327 KQ562145 KXJ76577.1 CP012525 ALC44610.1 GL442815 EFN62994.1 JXUM01137733 KQ568620 KXJ68944.1 KZ288451 PBC25573.1 GEDC01019705 JAS17593.1 JXJN01004400 CH477518 EAT39638.1 CH379069 EDY73389.2 KQ434878 KZC09901.1 CH902618 EDV39196.1 CM000159 EDW94523.1 GL767674 EFZ13246.1 NNAY01002176 OXU21868.1 GDIP01146506 GDIQ01203523 LRGB01000024 JAJ76896.1 JAK48202.1 KZS21683.1 GDIP01061564 JAM42151.1 GDIQ01048233 JAN46504.1 ADTU01008608 CH933809 KRG06649.1 GAMC01001475 JAC05081.1 EDW19480.1 KQ976453 KYM85324.1 KQ971321 EEZ99511.2 AXCN02001090 CH916366 EDV97318.1 CH940647 EDW70242.2 AAAB01008986 EAU75852.2 GAKP01001678 JAC57274.1 GDHF01007773 JAI44541.1 KQ979480 KYN21264.1 AAZO01005737 DS235816 EEB17576.1 OUUW01000002 SPP76054.1 KQ977513 KYN02182.1 APCN01002416 AXCM01009764 KQ982708 KYQ51803.1 JRES01000502 KNC30606.1 ATLV01023941 KE525347 KFB50085.1 GL888206 EGI65192.1 GDIQ01016728 JAN78009.1 GL732525 EFX88635.1 GDIQ01171333 JAK80392.1 CH954178 EDV51778.1 GDIP01030112 JAM73603.1 GEZM01062527 GEZM01062524 GEZM01062523 GEZM01062520 JAV69627.1 GBXI01012014 JAD02278.1 GEBQ01008409 JAT31568.1 GBHO01000591 JAG43013.1 GBRD01011958 GDHC01002291 JAG53866.1 JAQ16338.1 PZQS01000004 PVD32238.1 GGFK01012401 MBW45722.1 GGFL01004812 MBW68990.1 KQ414815 KOC60519.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000037510
+ More
UP000027135 UP000092462 UP000002320 UP000000304 UP000192223 UP000001292 UP000235965 UP000092461 UP000053097 UP000279307 UP000078200 UP000000803 UP000092444 UP000092445 UP000069940 UP000249989 UP000092553 UP000000311 UP000242457 UP000005203 UP000095300 UP000092460 UP000008820 UP000001819 UP000076502 UP000075920 UP000007801 UP000192221 UP000002282 UP000092443 UP000075880 UP000215335 UP000076858 UP000005205 UP000095301 UP000009192 UP000075901 UP000078540 UP000007266 UP000002358 UP000075886 UP000001070 UP000008792 UP000007062 UP000078492 UP000075882 UP000009046 UP000268350 UP000078542 UP000075840 UP000075883 UP000075902 UP000075809 UP000037069 UP000075903 UP000075900 UP000030765 UP000007755 UP000076407 UP000075881 UP000000305 UP000075884 UP000008711 UP000245119 UP000053825
UP000027135 UP000092462 UP000002320 UP000000304 UP000192223 UP000001292 UP000235965 UP000092461 UP000053097 UP000279307 UP000078200 UP000000803 UP000092444 UP000092445 UP000069940 UP000249989 UP000092553 UP000000311 UP000242457 UP000005203 UP000095300 UP000092460 UP000008820 UP000001819 UP000076502 UP000075920 UP000007801 UP000192221 UP000002282 UP000092443 UP000075880 UP000215335 UP000076858 UP000005205 UP000095301 UP000009192 UP000075901 UP000078540 UP000007266 UP000002358 UP000075886 UP000001070 UP000008792 UP000007062 UP000078492 UP000075882 UP000009046 UP000268350 UP000078542 UP000075840 UP000075883 UP000075902 UP000075809 UP000037069 UP000075903 UP000075900 UP000030765 UP000007755 UP000076407 UP000075881 UP000000305 UP000075884 UP000008711 UP000245119 UP000053825
Pfam
PF01974 tRNA_int_endo
Interpro
SUPFAM
SSF53032
SSF53032
Gene 3D
ProteinModelPortal
H9JQG9
A0A2H1WEF9
A0A2A4K666
A0A194PVP3
A0A1E1W6P0
A0A194QNP7
+ More
S4PDN4 A0A3S2N524 A0A0L7LV47 A0A067QI60 A0A1B0DJ44 B0X2T9 B4QK26 A0A1W4XHE4 B4HHS9 A0A2J7RA58 A0A2J7RA46 A0A1B0CSR0 A0A026W1B4 A0A1Q3F5L3 A0A1A9VFD4 Q0E8E7 D2NUI2 G4LU02 A0A1B0G9Y8 A0A1B0A2A3 A0A182GG98 A0A0M4EM16 E2AUD7 A0A182G1R5 A0A1S4FJZ8 A0A2A3E1E7 A0A088AVZ8 A0A1B6CWN3 A0A1I8P9N7 A0A1I8P9W0 A0A1B0AVW0 Q16YD2 B5DQS3 A0A154PDJ4 A0A182WDX8 B3MAW2 A0A1W4UKM1 B4PIG6 A0A1A9XR90 A0A1W4U8S9 E9J1N6 A0A182IQJ8 A0A232EU37 A0A0N8B842 A0A0P5Y840 A0A0P6FM94 A0A158P3X0 A0A1I8MDV3 A0A0Q9XG26 W8BUP8 A0A182TA15 B4KVK9 A0A195BKM6 D6WIR4 K7IQN6 A0A182Q2X7 B4J3T7 B4LF54 A0NGQ6 A0A034WRL9 A0A0K8W0B3 A0A151J8Z8 A0A182L517 E0VW20 A0A3B0J386 A0A195CNX8 A0A182HHI2 A0A182MHV5 A0A182UAP6 A0A151WVB4 A0A0L0CED2 A0A182VGL5 A0A182RRZ6 A0A084WIP1 F4WKV0 A0A182XTJ4 A0A182JRB0 A0A0P6HN96 E9FVZ2 A0A0P5LIF7 A0A182N2W8 B3ND56 A0A0P6AJ82 A0A1Y1LF38 A0A0A1WV44 A0A1B6M6M6 A0A0A9ZH52 A0A0K8SKM5 A0A2T7PFP2 A0A2M4AYD3 A0A2M4CUJ4 A0A2K6VAZ7 A0A0L7QPN5
S4PDN4 A0A3S2N524 A0A0L7LV47 A0A067QI60 A0A1B0DJ44 B0X2T9 B4QK26 A0A1W4XHE4 B4HHS9 A0A2J7RA58 A0A2J7RA46 A0A1B0CSR0 A0A026W1B4 A0A1Q3F5L3 A0A1A9VFD4 Q0E8E7 D2NUI2 G4LU02 A0A1B0G9Y8 A0A1B0A2A3 A0A182GG98 A0A0M4EM16 E2AUD7 A0A182G1R5 A0A1S4FJZ8 A0A2A3E1E7 A0A088AVZ8 A0A1B6CWN3 A0A1I8P9N7 A0A1I8P9W0 A0A1B0AVW0 Q16YD2 B5DQS3 A0A154PDJ4 A0A182WDX8 B3MAW2 A0A1W4UKM1 B4PIG6 A0A1A9XR90 A0A1W4U8S9 E9J1N6 A0A182IQJ8 A0A232EU37 A0A0N8B842 A0A0P5Y840 A0A0P6FM94 A0A158P3X0 A0A1I8MDV3 A0A0Q9XG26 W8BUP8 A0A182TA15 B4KVK9 A0A195BKM6 D6WIR4 K7IQN6 A0A182Q2X7 B4J3T7 B4LF54 A0NGQ6 A0A034WRL9 A0A0K8W0B3 A0A151J8Z8 A0A182L517 E0VW20 A0A3B0J386 A0A195CNX8 A0A182HHI2 A0A182MHV5 A0A182UAP6 A0A151WVB4 A0A0L0CED2 A0A182VGL5 A0A182RRZ6 A0A084WIP1 F4WKV0 A0A182XTJ4 A0A182JRB0 A0A0P6HN96 E9FVZ2 A0A0P5LIF7 A0A182N2W8 B3ND56 A0A0P6AJ82 A0A1Y1LF38 A0A0A1WV44 A0A1B6M6M6 A0A0A9ZH52 A0A0K8SKM5 A0A2T7PFP2 A0A2M4AYD3 A0A2M4CUJ4 A0A2K6VAZ7 A0A0L7QPN5
PDB
3P1Z
E-value=7.09754e-09,
Score=142
Ontologies
GO
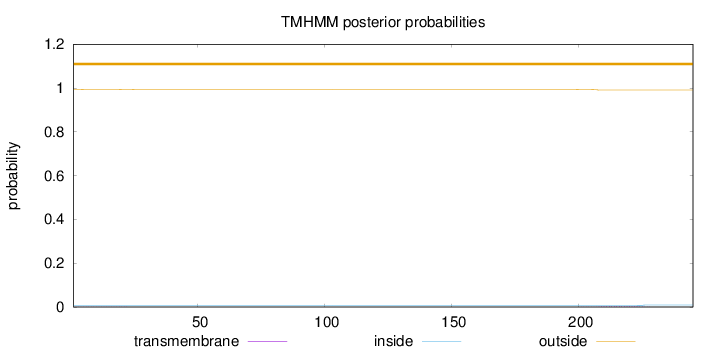
Topology
Length:
245
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05518
Exp number, first 60 AAs:
0.00723
Total prob of N-in:
0.00713
outside
1 - 245
Population Genetic Test Statistics
Pi
207.148638
Theta
166.683309
Tajima's D
0.693257
CLR
0
CSRT
0.572121393930303
Interpretation
Uncertain
