Gene
KWMTBOMO06653
Pre Gene Modal
BGIBMGA011777
Annotation
acidic_lipase_[Helicoverpa_armigera]
Full name
Lipase
Location in the cell
PlasmaMembrane Reliability : 2.899
Sequence
CDS
ATGCGATTTCAAGGCATACTTTTAATTTGTATGTTCTGTTTTAGTTTCGGAGATGCTCGATCATCTCCGAACGTTGAAGTGGTGAGAACACTTTTTGGTAACGACAAAGGCGCCCGTTATTCTAATAATGTAATCGAAGATGCTGAATTGGATGTGCCCGACCTAATCAGGAAATATGGCTATCCCGTAGAAGTTCATCACGTGTCCACCCCAGATGGTTATATCCTGGAAATGCATCGTATCCCTCATGGACGATATGCGAACAACAGACCTAATAGTAAAAAACCTGTCGTTTTCCTGATGCACGGTCTGCTCTGTTCCTCAGCTGACTGGGTGATGACAGGCCCTAAACTTGGACTGGGTTATATTCTAGCTGAAGAGGGATATGATGTTTGGATGGGTAACGCTAGAGGCAATTATTATTCCCGAAGACACATTTCTTTAAACCCAGATAGCTTCATTAGTACTCAATTTTGGAAATTTTCTTGGGACGAGATAGGAAATATAGATTTGCCCGCTATGATAGATTTTTCACTGAGGAGAAGTGGACAAAAAAAATTGCATTATATTGGGCATTCTCAGGGATCAACTGCCTTCTTTGTGATGACATCATTACGTCCGGAGTATAACCAGAAAATCATATCAATGCAAGCGTTGGCACCTGTAGCGTACATGGGTCACAACAAAAATCCTTTATTGCTAGCTATAGCACCATTCGCCCATGAATTAGTGACTTTAGCATCATTAATAGGAATTGGGGAGTTCTTGCCGAACAGTAGGGTTTTCACTTGGGCAGGACAAACCTTTTGTAAGGATGAAGTTGTGTTTCAACCTATATGCAGCAACATTCTTTTCTTGATCGGTGGATGGAACGAACAACAGCTCAACACGACTTTACTGCCCGTGAAATTCGGTCACACTCCCGCTGGTGCTGCAGCAAGACAGTTGGCTCATTATGGTCAAGGAATTGCCGGAAAGGAATTCCGTCGTTACGACCACGGTAGTAGACTAGCAAACAGAAGAGCCTATGGAACTAGCACTCCGCCAAAGTACGACTTATCGAAAGTGACCGCACCCGTGTTCCTTCACTATTCCGACAATGATCCTCTCGCCGAAATCGAGGATGTTGAGATATTGTATAGAGAGCTCGGAAACCCAGTAGGAATGTTCCGGGTGCCCTTGGCGACATTCAACCATTTAGATTTCTTGTGGGCCATTGACGTTAGGACTTTGTTATACGACACTGTCATCTATTATATGAAAACAATGAACGAGGACGCTTAA
Protein
MRFQGILLICMFCFSFGDARSSPNVEVVRTLFGNDKGARYSNNVIEDAELDVPDLIRKYGYPVEVHHVSTPDGYILEMHRIPHGRYANNRPNSKKPVVFLMHGLLCSSADWVMTGPKLGLGYILAEEGYDVWMGNARGNYYSRRHISLNPDSFISTQFWKFSWDEIGNIDLPAMIDFSLRRSGQKKLHYIGHSQGSTAFFVMTSLRPEYNQKIISMQALAPVAYMGHNKNPLLLAIAPFAHELVTLASLIGIGEFLPNSRVFTWAGQTFCKDEVVFQPICSNILFLIGGWNEQQLNTTLLPVKFGHTPAGAAARQLAHYGQGIAGKEFRRYDHGSRLANRRAYGTSTPPKYDLSKVTAPVFLHYSDNDPLAEIEDVEILYRELGNPVGMFRVPLATFNHLDFLWAIDVRTLLYDTVIYYMKTMNEDA
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9JQH1
A0A2A4K5V7
A0A2A4K5R5
I3QC10
A0A2H1VT83
A0A2H1WEG8
+ More
A0A2A4K660 A0A194PXA6 A0A3S2NJM8 A0A194QUC1 A0A2A4K6W6 A0A2H1WZN4 A0A194QQB7 A0A194PWK6 A0A194Q1W6 A0A194QNR0 A0A2A4K595 A0A194QNQ2 A0A194PVH3 A0A3S2LGI4 H9JM87 A0A194PVN8 H9JM86 A0A1E1W7Q1 A0A212FG21 A0A212F4Q9 A0A0N0PG73 H9J831 A0A2A4IVE2 A0A1L8DPY1 A0A1L8DPM5 A0A212FB51 A0A1L8DQ28 A0A1L8DPS2 A0A194R2Y3 A0A1L8DQ21 A0A3S2NF67 A0A2H1X0E5 A0A194PL00 A0A3S2P1G5 A0A1L8DPQ4 A0A1L8DPW0 A0A084VDU8 A0A182QBZ9 A0A1Y9H2S0 Q16MC6 A0A2A4JD47 A0A1L8DYQ8 I3QC09 A0A2H1X102 A0A067RCF7 A0A182R887 W5J6X8 A0A182J854 A0A182U574 A0A182UUQ4 A0A182KX90 A0A1I8JTB7 A0A0T6BEX7 Q5TVS6 A0A1Y9J0B7 A0A182JMM3 T1EA03 A0A1L8DZ68 A0A084W6W2 A0A139WJS1 A0A182P083 I1SR89 D2A0X3 B0W6G8 A0A182KCP4 A0A1L8DYT2 A0A182K065 Q16F27 A0A1B0DL47 A0A182TPG6 A0A182H3N2 Q16MD1 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182ML09 A0A182UTW8 A0A182V7K7 A0A1S4FVZ7 A0A026W0L2 A0A1Y9J0S8 A0A182KX99 A0A1I8JTB8 A0A182YLM4 B0W6G4 D2A0X6 A0A0J7NNT2 A0A1B0CTU8 A0A182FL00 A0A1Y9H2R7 A0A2J7QW32 Q7QH37 W5JF51
A0A2A4K660 A0A194PXA6 A0A3S2NJM8 A0A194QUC1 A0A2A4K6W6 A0A2H1WZN4 A0A194QQB7 A0A194PWK6 A0A194Q1W6 A0A194QNR0 A0A2A4K595 A0A194QNQ2 A0A194PVH3 A0A3S2LGI4 H9JM87 A0A194PVN8 H9JM86 A0A1E1W7Q1 A0A212FG21 A0A212F4Q9 A0A0N0PG73 H9J831 A0A2A4IVE2 A0A1L8DPY1 A0A1L8DPM5 A0A212FB51 A0A1L8DQ28 A0A1L8DPS2 A0A194R2Y3 A0A1L8DQ21 A0A3S2NF67 A0A2H1X0E5 A0A194PL00 A0A3S2P1G5 A0A1L8DPQ4 A0A1L8DPW0 A0A084VDU8 A0A182QBZ9 A0A1Y9H2S0 Q16MC6 A0A2A4JD47 A0A1L8DYQ8 I3QC09 A0A2H1X102 A0A067RCF7 A0A182R887 W5J6X8 A0A182J854 A0A182U574 A0A182UUQ4 A0A182KX90 A0A1I8JTB7 A0A0T6BEX7 Q5TVS6 A0A1Y9J0B7 A0A182JMM3 T1EA03 A0A1L8DZ68 A0A084W6W2 A0A139WJS1 A0A182P083 I1SR89 D2A0X3 B0W6G8 A0A182KCP4 A0A1L8DYT2 A0A182K065 Q16F27 A0A1B0DL47 A0A182TPG6 A0A182H3N2 Q16MD1 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182ML09 A0A182UTW8 A0A182V7K7 A0A1S4FVZ7 A0A026W0L2 A0A1Y9J0S8 A0A182KX99 A0A1I8JTB8 A0A182YLM4 B0W6G4 D2A0X6 A0A0J7NNT2 A0A1B0CTU8 A0A182FL00 A0A1Y9H2R7 A0A2J7QW32 Q7QH37 W5JF51
Pubmed
EMBL
BABH01011980
NWSH01000114
PCG79406.1
PCG79409.1
JF999959
AFI64313.1
+ More
ODYU01004000 SOQ43434.1 ODYU01008119 SOQ51451.1 PCG79408.1 KQ459590 KPI97394.1 RSAL01000448 RVE41683.1 KQ461190 KPJ07151.1 PCG79412.1 ODYU01012265 SOQ58477.1 KPJ07150.1 KPI97393.1 KPI97395.1 KPJ07152.1 PCG79407.1 KPJ07153.1 KPI97391.1 RSAL01000137 RVE46199.1 BABH01033349 BABH01033350 BABH01033351 BABH01033352 KPI97392.1 BABH01033355 GDQN01008173 JAT82881.1 AGBW02008724 OWR52685.1 AGBW02010324 OWR48717.1 LADJ01067081 KPJ21692.1 BABH01020326 NWSH01005801 PCG63937.1 GFDF01005647 JAV08437.1 GFDF01005646 JAV08438.1 AGBW02009374 OWR50972.1 GFDF01005644 JAV08440.1 GFDF01005645 JAV08439.1 KQ460855 KPJ11879.1 GFDF01005639 JAV08445.1 RSAL01000145 RVE45945.1 ODYU01012460 SOQ58781.1 KQ459601 KPI94007.1 RSAL01000004 RVE54656.1 GFDF01005642 JAV08442.1 GFDF01005643 JAV08441.1 ATLV01011799 KE524723 KFB36142.1 AXCN02000090 CH477866 EAT35491.1 NWSH01001918 PCG69686.1 GFDF01002510 JAV11574.1 JF999958 AFI64312.1 ODYU01012554 SOQ58947.1 KK852794 KDR16428.1 ADMH02002130 ETN58534.1 APCN01002227 LJIG01001091 KRT85849.1 AAAB01008820 EAL41497.4 GAMD01001453 JAB00138.1 GFDF01002509 JAV11575.1 ATLV01020990 KE525311 KFB45956.1 KQ971338 KYB28091.1 JF303742 ADZ54058.1 EFA01616.1 DS231848 EDS36642.1 GFDF01002505 JAV11579.1 CH478517 EAT32843.1 AJVK01016188 JXUM01107700 KQ565165 KXJ71265.1 EAT35487.1 APCN01000436 AAAB01008859 EAA08216.4 AXCM01003911 KK107599 QOIP01000012 EZA48594.1 RLU15932.1 EDS36638.1 EFA01613.1 LBMM01002941 KMQ94155.1 AJWK01028000 NEVH01009768 PNF32784.1 EAA05393.4 ADMH02001587 ETN61958.1
ODYU01004000 SOQ43434.1 ODYU01008119 SOQ51451.1 PCG79408.1 KQ459590 KPI97394.1 RSAL01000448 RVE41683.1 KQ461190 KPJ07151.1 PCG79412.1 ODYU01012265 SOQ58477.1 KPJ07150.1 KPI97393.1 KPI97395.1 KPJ07152.1 PCG79407.1 KPJ07153.1 KPI97391.1 RSAL01000137 RVE46199.1 BABH01033349 BABH01033350 BABH01033351 BABH01033352 KPI97392.1 BABH01033355 GDQN01008173 JAT82881.1 AGBW02008724 OWR52685.1 AGBW02010324 OWR48717.1 LADJ01067081 KPJ21692.1 BABH01020326 NWSH01005801 PCG63937.1 GFDF01005647 JAV08437.1 GFDF01005646 JAV08438.1 AGBW02009374 OWR50972.1 GFDF01005644 JAV08440.1 GFDF01005645 JAV08439.1 KQ460855 KPJ11879.1 GFDF01005639 JAV08445.1 RSAL01000145 RVE45945.1 ODYU01012460 SOQ58781.1 KQ459601 KPI94007.1 RSAL01000004 RVE54656.1 GFDF01005642 JAV08442.1 GFDF01005643 JAV08441.1 ATLV01011799 KE524723 KFB36142.1 AXCN02000090 CH477866 EAT35491.1 NWSH01001918 PCG69686.1 GFDF01002510 JAV11574.1 JF999958 AFI64312.1 ODYU01012554 SOQ58947.1 KK852794 KDR16428.1 ADMH02002130 ETN58534.1 APCN01002227 LJIG01001091 KRT85849.1 AAAB01008820 EAL41497.4 GAMD01001453 JAB00138.1 GFDF01002509 JAV11575.1 ATLV01020990 KE525311 KFB45956.1 KQ971338 KYB28091.1 JF303742 ADZ54058.1 EFA01616.1 DS231848 EDS36642.1 GFDF01002505 JAV11579.1 CH478517 EAT32843.1 AJVK01016188 JXUM01107700 KQ565165 KXJ71265.1 EAT35487.1 APCN01000436 AAAB01008859 EAA08216.4 AXCM01003911 KK107599 QOIP01000012 EZA48594.1 RLU15932.1 EDS36638.1 EFA01613.1 LBMM01002941 KMQ94155.1 AJWK01028000 NEVH01009768 PNF32784.1 EAA05393.4 ADMH02001587 ETN61958.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000007151
+ More
UP000030765 UP000075886 UP000075884 UP000008820 UP000027135 UP000075900 UP000000673 UP000075880 UP000075902 UP000075903 UP000075882 UP000075840 UP000007062 UP000076407 UP000007266 UP000075885 UP000002320 UP000075881 UP000092462 UP000069940 UP000249989 UP000075883 UP000053097 UP000279307 UP000076408 UP000036403 UP000092461 UP000069272 UP000235965
UP000030765 UP000075886 UP000075884 UP000008820 UP000027135 UP000075900 UP000000673 UP000075880 UP000075902 UP000075903 UP000075882 UP000075840 UP000007062 UP000076407 UP000007266 UP000075885 UP000002320 UP000075881 UP000092462 UP000069940 UP000249989 UP000075883 UP000053097 UP000279307 UP000076408 UP000036403 UP000092461 UP000069272 UP000235965
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JQH1
A0A2A4K5V7
A0A2A4K5R5
I3QC10
A0A2H1VT83
A0A2H1WEG8
+ More
A0A2A4K660 A0A194PXA6 A0A3S2NJM8 A0A194QUC1 A0A2A4K6W6 A0A2H1WZN4 A0A194QQB7 A0A194PWK6 A0A194Q1W6 A0A194QNR0 A0A2A4K595 A0A194QNQ2 A0A194PVH3 A0A3S2LGI4 H9JM87 A0A194PVN8 H9JM86 A0A1E1W7Q1 A0A212FG21 A0A212F4Q9 A0A0N0PG73 H9J831 A0A2A4IVE2 A0A1L8DPY1 A0A1L8DPM5 A0A212FB51 A0A1L8DQ28 A0A1L8DPS2 A0A194R2Y3 A0A1L8DQ21 A0A3S2NF67 A0A2H1X0E5 A0A194PL00 A0A3S2P1G5 A0A1L8DPQ4 A0A1L8DPW0 A0A084VDU8 A0A182QBZ9 A0A1Y9H2S0 Q16MC6 A0A2A4JD47 A0A1L8DYQ8 I3QC09 A0A2H1X102 A0A067RCF7 A0A182R887 W5J6X8 A0A182J854 A0A182U574 A0A182UUQ4 A0A182KX90 A0A1I8JTB7 A0A0T6BEX7 Q5TVS6 A0A1Y9J0B7 A0A182JMM3 T1EA03 A0A1L8DZ68 A0A084W6W2 A0A139WJS1 A0A182P083 I1SR89 D2A0X3 B0W6G8 A0A182KCP4 A0A1L8DYT2 A0A182K065 Q16F27 A0A1B0DL47 A0A182TPG6 A0A182H3N2 Q16MD1 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182ML09 A0A182UTW8 A0A182V7K7 A0A1S4FVZ7 A0A026W0L2 A0A1Y9J0S8 A0A182KX99 A0A1I8JTB8 A0A182YLM4 B0W6G4 D2A0X6 A0A0J7NNT2 A0A1B0CTU8 A0A182FL00 A0A1Y9H2R7 A0A2J7QW32 Q7QH37 W5JF51
A0A2A4K660 A0A194PXA6 A0A3S2NJM8 A0A194QUC1 A0A2A4K6W6 A0A2H1WZN4 A0A194QQB7 A0A194PWK6 A0A194Q1W6 A0A194QNR0 A0A2A4K595 A0A194QNQ2 A0A194PVH3 A0A3S2LGI4 H9JM87 A0A194PVN8 H9JM86 A0A1E1W7Q1 A0A212FG21 A0A212F4Q9 A0A0N0PG73 H9J831 A0A2A4IVE2 A0A1L8DPY1 A0A1L8DPM5 A0A212FB51 A0A1L8DQ28 A0A1L8DPS2 A0A194R2Y3 A0A1L8DQ21 A0A3S2NF67 A0A2H1X0E5 A0A194PL00 A0A3S2P1G5 A0A1L8DPQ4 A0A1L8DPW0 A0A084VDU8 A0A182QBZ9 A0A1Y9H2S0 Q16MC6 A0A2A4JD47 A0A1L8DYQ8 I3QC09 A0A2H1X102 A0A067RCF7 A0A182R887 W5J6X8 A0A182J854 A0A182U574 A0A182UUQ4 A0A182KX90 A0A1I8JTB7 A0A0T6BEX7 Q5TVS6 A0A1Y9J0B7 A0A182JMM3 T1EA03 A0A1L8DZ68 A0A084W6W2 A0A139WJS1 A0A182P083 I1SR89 D2A0X3 B0W6G8 A0A182KCP4 A0A1L8DYT2 A0A182K065 Q16F27 A0A1B0DL47 A0A182TPG6 A0A182H3N2 Q16MD1 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182ML09 A0A182UTW8 A0A182V7K7 A0A1S4FVZ7 A0A026W0L2 A0A1Y9J0S8 A0A182KX99 A0A1I8JTB8 A0A182YLM4 B0W6G4 D2A0X6 A0A0J7NNT2 A0A1B0CTU8 A0A182FL00 A0A1Y9H2R7 A0A2J7QW32 Q7QH37 W5JF51
PDB
1K8Q
E-value=1.17409e-72,
Score=695
Ontologies
GO
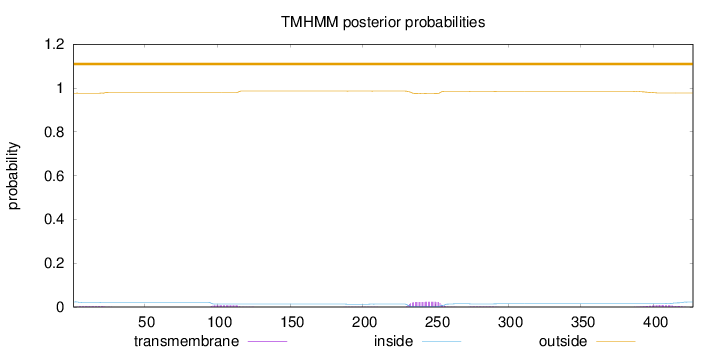
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.997256
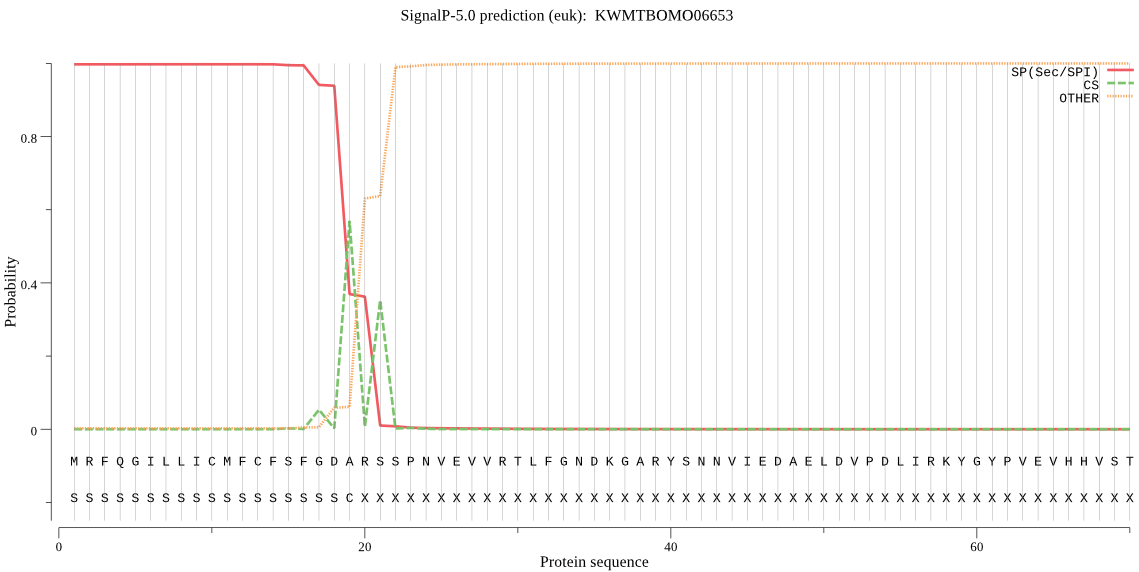
Length:
427
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.93806
Exp number, first 60 AAs:
0.06176
Total prob of N-in:
0.02340
outside
1 - 427
Population Genetic Test Statistics
Pi
248.091205
Theta
227.407307
Tajima's D
0.240775
CLR
0.400494
CSRT
0.441277936103195
Interpretation
Uncertain
