Gene
KWMTBOMO06651
Pre Gene Modal
BGIBMGA011704
Annotation
PREDICTED:_BAI1-associated_protein_3_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.502 Nuclear Reliability : 1.93
Sequence
CDS
ATGAGTTTCTTAACATCATTGGGCCAGTATGTGGCGGGCGTAGCTGACAGTGTAGCCGGTCTGGCGCTCTCACCGCGGCGTGCACCGCCCTCACGACATCGCTCTGCCGAGGAGGGTCAGGCACAACAACCTCCACCGTCAACCGCTACTATGCGTGGATTTCCTAGAGTTCTACCAACAGAAGGCGCTTCTGCCCTGGGCGCCCGACGGCGCACTCTAGATTGCTTGGCTCCAGCACCAGCTCAACGACGCGGTGGATCACTCCGGGTGCCCCGTCACCAAGCACCACCTGAAAAACCCACGATGGCATTCTGCAAAAGACGTCTCTCTTGGCCAGAGGTCGATAACTCTGTGAACTCGGGGGTTCAAGACACGGATGGCTCTTACTTCGAAAGTTTCACGGCGCTGTCTTGGCGACAGGAGAACAGGCGGCTAGCAGCATTGAGACTGGCCGAAGCTAGGGCGGAACGCCCACCACCCATGCCCCAAGACCTTGGCCTGCCAAGTGTGTCTGCGCAACTTTCTCATAAAGAACATGAGCATTTGTACACTGAGGTACTATACTGCATTGAGAACGCAGTGGGAGCTCCGGCTCCAGACGGCCAATTCGCTGCTTACAAAGAAGAAGTGATAGAATACGCGAGGCGTGCTTTCCGAATTTCACAAGAAGTTCACGCGCGTGCTATCCGCGCCGCCGGCAGTGAGCGACCACCTACTGTCGTGCTCGGCTGCTCTGTCATAGAAGCTGATGGACTAGAGGCCAAGGATGCCAATGGGTTCAGCGATCCATATTGCATGTTGGGGATTCAACCAGCTTCTTTACCGGCATCACCTCGTGCATCAGAGGACGAAGGCCGCAAGCACGGGTTCCGTCTCAGCTTCAAGCGAAGAGACGGTCGACAACAACGAGACAGTCTTGGTAGACTCCCGGCACGTTACATCCGAGCCACGAGCGTACGACCACATACGCTCTCACCGAAATGGAATGAAACCTTCAAATTTGATATTGATGACATCTCATCGGATGTACTGCATCTGGACATTTGGGACCACGACGACGAGAGCTCCGTACTAGATGCTGTCTCTCGGCTGAACGAGGTCCGTGGAGTAAGGGGTCTTGGCCGCTTCTTTAAGCAAGTGTGTCAAAGCGCACGACAGGGAAGCCAGGATGACTTTCTAGGATGTGTGAACATACCTCTGCGGGAGATTCCGTCGACTGGCATCGAAGCTTGGTACAAGTTGGAGGCACGCTCACAACGCTCTTCGGTTCAAGGACGAATCCGCTTACGCCTCTGGTTATCAGCTCGTGAGGCAGGCCGCCACGACGACGACAATTGGCAACAGGTTCGTCAACACGAGCGCTTGTTCGGGGTTTTACTGTCGTATGAAGTGGACACGGCGGGTATGGTGAACGTAGTCGAGGGCGAAGCGAGCTCTGAGTGGGAAGGTGACTTATGCGGGGCAGCTCAAACTCTTTTGCATCAACATGCTATACAAGGAGACATGAGCGCGCTGCAAGCCGCCATCGCCCGTTTCGCTGCGGCTTGTAAACTGAACTGCGAAGCTCCTCTCGATCCCAAGTACATGTACAAATTGTTGACGGAGCTCGAACGAGCCTGGTGTGCGAGCGAGGCGGAGGTCGGCAGTTGCGCCGGGGAAAGCGCGTCGGGCCTCTCCCGCGACGAGGAACGCTGGCTCGCCGACTGCTTCGGAGAGTTTGTCGAACGCGGCCTGCATCAGATGCGACTCCACCGGGACCTCTATCCGGTGCTCCACGGCAATAGCTTGAACAAGTTGGAGTATCTACTGCGGTGTCTCAGCCAGCTGTCACAGATGAAAGCGTTTTGGCGTTGCTGTCCCTTCAATAAGGAAATCAGGAGTGAAATAGTGAGTTCCCTTCGCAAAGGCACTTCGGAATGGTACGAAGCGCTCTGTGCACAAGACACTGAGTCCGAGCGTGGCGAAGATCTTGTGAACCTCATCACACTAGTACAAATAGATCTTCAACAGGGGCTGACCTATTATCATCCGTTGTTCGAGATGACAAACAATGTTCCGTACTTTAGCGTGGTTTTCAATCAGATCGACAAAATGGTGACGGAAGACGTAAGGCAATGCGTCGAGCACTGGGTGGGTGAAGGTTGGTGCGGTGTGGCTGAGGAAGAATTATATGACGAACATCAGAATCCCATGTGCATAGTATCAGCAAAGACGCTGCCATTCGAGGTGTTGTCAGCGGTACGAGAGCTGTGCGCTGCAGCCGCCCCTGCGCCGCACTCGCCACACGATCCACCACCGCACCTGTTACATGCGTATTTATGGTTCGAGCCACTCATCGATCGTTGGCTGGCCGTCACCAAAACAATGGCGCTACAGCGCGTGCGAGCCGCCGTCGATCTGGATCGTCCGGTTGAAGGGGAACATCTCGTGAAGCATAGTACTTCATCAGTGGACACCGCAGCTTGTCTTTATCACATGAGATTGGTCTGGCGAGCTATTGGTGGAGCCGAGGAAGGTACTACTGGGGGCTCGCTGAGACTCCTTGAGGCAGCCTGTGCCACTGCACTCCACTACACCGACTTAGTCCACGGACAACTTGCCGATCAGGGATATTACGAGAACCACGGTCAAAACAAAACAACCGAGGAAATATGCACGATCGTGAATAATCTGGAGTACGTCCGACGCTCGCTCTCTGAGTACGACGATGAACACATTGCCAGCGATGAGAACTCCCGTCAGCTAGTGCGCGGTGCTCTGGGTACTCTGGACGCGCGATCCGCTCGTGCTGCCGCCCCGCTGGCCGCAAGAGCACGGCCACCTCTACGCAAGGCCGTGTTCCATCTAGCGTGGTCACCTGACACGCTACCTACCATACAGGCCATAACTCCACTTTTGGAGTTCCTAGACCAGCACCTGTCAGCGTTGAATACATGGCTGCTTCCCCGTGCCTTCCTCCGAGCTCTTGCTGGTTGCTGGAGCGCCGTACTCAAGGAAGTGTCAACACAAGCTGATGCTGGAGGCGCTGAGAGACCTAGGGTGTACCATCACAGACTGAGAGAGGCTCTTGACTTACTCGCCGAGTTCTTCCACGCTGAAGGAAAAGGTCTCCCGATGACAGACGTCCACAACTTGGAATGGGGTCGCGTAGAGCAACGCATGCAGTATCATCAGGCACCGACTGAGGAGTTGCTCGAATTATGGCTCGCCGATAGACTGGCTGAGCAGGCTCGGACACCAATGCCTTCGCCATATGGATCTATACTTGTCAGAGTGTATTTCAACCACGACTCGCTCTGCGTGGAGGTGTTGTCCGCGCGTGACGTGATCCCCCTGGACCCGAACGGCCTCAGTGATCCGTTCGTAGTGCTCGAACTTCTTCCGAAGCGACTCTTCCCTAAAGCCCACGAACAAAGCACAAATGTTCAGAAGAAAACTCTGAACCCCGTGTGGGACGAGTGCTTCGAGTTCGCGGTGTCGTTGGAGTCGTGTCGGTCGTCGCAAGCGGCGCTCGCTCTGTCCGTGTGGGACCGGGACGTCTTGACTGCCGACGACTTCGCCGGTGAGGCGTTCATTCCGCTGTCGCGCGTCCCCGGGGTGCACACTCACGCGCCCCCCGACCCCCTGCGTCCCGTCGAGCTGCCCCTCATGCAGCTCCATGACAGGAATCATCCGATATTGCAGATACTGGAATCTCGCACCACCGATAAGTTCGCCATGGATTTCGTTAAAAAACAAAAATTGCGTTTTGCTGAACAGTAA
Protein
MSFLTSLGQYVAGVADSVAGLALSPRRAPPSRHRSAEEGQAQQPPPSTATMRGFPRVLPTEGASALGARRRTLDCLAPAPAQRRGGSLRVPRHQAPPEKPTMAFCKRRLSWPEVDNSVNSGVQDTDGSYFESFTALSWRQENRRLAALRLAEARAERPPPMPQDLGLPSVSAQLSHKEHEHLYTEVLYCIENAVGAPAPDGQFAAYKEEVIEYARRAFRISQEVHARAIRAAGSERPPTVVLGCSVIEADGLEAKDANGFSDPYCMLGIQPASLPASPRASEDEGRKHGFRLSFKRRDGRQQRDSLGRLPARYIRATSVRPHTLSPKWNETFKFDIDDISSDVLHLDIWDHDDESSVLDAVSRLNEVRGVRGLGRFFKQVCQSARQGSQDDFLGCVNIPLREIPSTGIEAWYKLEARSQRSSVQGRIRLRLWLSAREAGRHDDDNWQQVRQHERLFGVLLSYEVDTAGMVNVVEGEASSEWEGDLCGAAQTLLHQHAIQGDMSALQAAIARFAAACKLNCEAPLDPKYMYKLLTELERAWCASEAEVGSCAGESASGLSRDEERWLADCFGEFVERGLHQMRLHRDLYPVLHGNSLNKLEYLLRCLSQLSQMKAFWRCCPFNKEIRSEIVSSLRKGTSEWYEALCAQDTESERGEDLVNLITLVQIDLQQGLTYYHPLFEMTNNVPYFSVVFNQIDKMVTEDVRQCVEHWVGEGWCGVAEEELYDEHQNPMCIVSAKTLPFEVLSAVRELCAAAAPAPHSPHDPPPHLLHAYLWFEPLIDRWLAVTKTMALQRVRAAVDLDRPVEGEHLVKHSTSSVDTAACLYHMRLVWRAIGGAEEGTTGGSLRLLEAACATALHYTDLVHGQLADQGYYENHGQNKTTEEICTIVNNLEYVRRSLSEYDDEHIASDENSRQLVRGALGTLDARSARAAAPLAARARPPLRKAVFHLAWSPDTLPTIQAITPLLEFLDQHLSALNTWLLPRAFLRALAGCWSAVLKEVSTQADAGGAERPRVYHHRLREALDLLAEFFHAEGKGLPMTDVHNLEWGRVEQRMQYHQAPTEELLELWLADRLAEQARTPMPSPYGSILVRVYFNHDSLCVEVLSARDVIPLDPNGLSDPFVVLELLPKRLFPKAHEQSTNVQKKTLNPVWDECFEFAVSLESCRSSQAALALSVWDRDVLTADDFAGEAFIPLSRVPGVHTHAPPDPLRPVELPLMQLHDRNHPILQILESRTTDKFAMDFVKKQKLRFAEQ
Summary
Uniprot
A0A2A4K676
A0A194Q1W1
A0A194QP76
D6WGN2
N6TXN5
A0A1B0G330
+ More
A0A1A9ZF16 A0A1A9V8Y9 B4J1Z0 A0A1A9Y5I4 A0A0L0CEF3 Q29E82 A0A3B0JUZ2 B4HV40 B4LC91 Q9VS41 V9ID48 B3NFG0 A0A1W4UGT6 B4PK49 A0A336M3V5 B4MLD3 B3M4Z2 A0A1J1IK11 A0A1I8PCN0 A0A195FTC2 A0A151WWG4 U4TYT9 A0A195DKD8 A0A0L7QN91 A0A195D7D9 A0A3L8E2W9 A0A195B3G4 A0A0J9RQE5 A0A158P192 A0A1A9W7M7 A0A1Y1KZ13 A0A067RCT7 A0A0M3QWM5 Q7Q6J0 B4L958 B4QKB5 A0A026WF15 E2BQS6 A0A0M8ZUB9 Q6NN63 A0A084WDE4 A7UTW6 A0A310SJL0 J9JVL3 A0A2H8TEI2 E9IBZ6 A0A182IWZ2 A0A182MIJ4 A0A182KQ24 A0A182V040 A0A182XMG4 A0A182FDW1 W5JM45 A0A182WL80 A0A182PIG1 A0A182QI95 A0A182NJH4 A0A182I294 A0A182YJA7 A0A2S2QF62 A0A182GVW5 A0A182TJ31 Q17HF2 A0A182S617 A0A182R468 A0A1B0BRC2 B0WI97 B4HAT0 A0A182K8M0 E0VGL7 A0A1I8MMB7 E9FTH5 A0A2J7R494 A0A1S3HNM7 A0A1S3HNN6 A0A0J7L115 A0A210QA78 A0A2T7Q0X1 V4BE93 A0A147BQR0 K1PNI3 R7UU63 A0A2R5LCG0 B7PPK2 A0A0K2T6Q5 W5M7U2 W5M7S9
A0A1A9ZF16 A0A1A9V8Y9 B4J1Z0 A0A1A9Y5I4 A0A0L0CEF3 Q29E82 A0A3B0JUZ2 B4HV40 B4LC91 Q9VS41 V9ID48 B3NFG0 A0A1W4UGT6 B4PK49 A0A336M3V5 B4MLD3 B3M4Z2 A0A1J1IK11 A0A1I8PCN0 A0A195FTC2 A0A151WWG4 U4TYT9 A0A195DKD8 A0A0L7QN91 A0A195D7D9 A0A3L8E2W9 A0A195B3G4 A0A0J9RQE5 A0A158P192 A0A1A9W7M7 A0A1Y1KZ13 A0A067RCT7 A0A0M3QWM5 Q7Q6J0 B4L958 B4QKB5 A0A026WF15 E2BQS6 A0A0M8ZUB9 Q6NN63 A0A084WDE4 A7UTW6 A0A310SJL0 J9JVL3 A0A2H8TEI2 E9IBZ6 A0A182IWZ2 A0A182MIJ4 A0A182KQ24 A0A182V040 A0A182XMG4 A0A182FDW1 W5JM45 A0A182WL80 A0A182PIG1 A0A182QI95 A0A182NJH4 A0A182I294 A0A182YJA7 A0A2S2QF62 A0A182GVW5 A0A182TJ31 Q17HF2 A0A182S617 A0A182R468 A0A1B0BRC2 B0WI97 B4HAT0 A0A182K8M0 E0VGL7 A0A1I8MMB7 E9FTH5 A0A2J7R494 A0A1S3HNM7 A0A1S3HNN6 A0A0J7L115 A0A210QA78 A0A2T7Q0X1 V4BE93 A0A147BQR0 K1PNI3 R7UU63 A0A2R5LCG0 B7PPK2 A0A0K2T6Q5 W5M7U2 W5M7S9
Pubmed
26354079
18362917
19820115
23537049
17994087
26108605
+ More
15632085 23185243 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 22936249 21347285 28004739 24845553 12364791 24508170 20798317 24438588 21282665 20966253 20920257 23761445 25244985 26483478 17510324 20566863 25315136 21292972 28812685 23254933 29652888 22992520
15632085 23185243 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 22936249 21347285 28004739 24845553 12364791 24508170 20798317 24438588 21282665 20966253 20920257 23761445 25244985 26483478 17510324 20566863 25315136 21292972 28812685 23254933 29652888 22992520
EMBL
NWSH01000114
PCG79414.1
KQ459590
KPI97390.1
KQ461190
KPJ07154.1
+ More
KQ971321 EFA00156.1 APGK01050981 APGK01050982 APGK01050983 APGK01050984 APGK01050985 APGK01050986 KB741180 ENN73131.1 CCAG010009843 CH916366 EDV95915.1 JRES01000493 KNC30788.1 CH379070 EAL30180.2 KRT08700.1 OUUW01000002 SPP77529.1 CH480817 EDW50811.1 CH940647 EDW68736.1 KRF83994.1 AE014296 AAF50588.4 AGB94199.1 JR038769 AEY58401.1 CH954178 EDV50502.1 CM000159 EDW93729.1 UFQT01000369 SSX23613.1 CH963847 EDW73391.2 CH902618 EDV40566.1 CVRI01000050 CRK99406.1 KQ981272 KYN43860.1 KQ982691 KYQ52210.1 KB631768 ERL85992.1 KQ980765 KYN13360.1 KQ414855 KOC60100.1 KQ976749 KYN08772.1 QOIP01000001 RLU26872.1 KQ976641 KYM78810.1 CM002912 KMY98048.1 ADTU01006076 ADTU01006077 ADTU01006078 ADTU01006079 ADTU01006080 GEZM01072759 JAV65410.1 KK852721 KDR17747.1 CP012525 ALC44409.1 AAAB01008960 EAA11102.4 CH933816 EDW17233.2 CM000363 EDX09538.1 KK107242 EZA54548.1 GL449799 EFN81951.1 KQ435841 KOX71305.1 BT011431 AAR99089.1 ATLV01022998 KE525339 KFB48238.1 EDO63740.1 KQ761906 OAD56453.1 ABLF02034728 ABLF02034739 ABLF02034740 ABLF02057270 GFXV01000680 MBW12485.1 GL762140 EFZ21915.1 AXCM01010389 ADMH02001139 ETN63965.1 AXCN02000446 APCN01000131 GGMS01007130 MBY76333.1 JXUM01091842 JXUM01091843 KQ563946 KXJ73031.1 CH477250 EAT46054.1 JXJN01019032 JXJN01019033 JXJN01019034 DS231945 EDS28323.1 CH479244 EDW37712.1 DS235149 EEB12523.1 GL732524 EFX89361.1 NEVH01007814 PNF35637.1 LBMM01001340 KMQ96482.1 NEDP02004422 OWF45641.1 PZQS01000001 PVD39324.1 KB202953 ESO87159.1 GEGO01002562 JAR92842.1 JH818302 EKC23238.1 AMQN01007080 KB299905 ELU07462.1 GGLE01003042 MBY07168.1 ABJB010047782 ABJB010056371 ABJB010060520 ABJB010093577 ABJB010181485 ABJB010222402 ABJB010363018 ABJB010448035 ABJB010514674 ABJB010582315 DS758678 EEC08524.1 HACA01004114 CDW21475.1 AHAT01029956 AHAT01029957
KQ971321 EFA00156.1 APGK01050981 APGK01050982 APGK01050983 APGK01050984 APGK01050985 APGK01050986 KB741180 ENN73131.1 CCAG010009843 CH916366 EDV95915.1 JRES01000493 KNC30788.1 CH379070 EAL30180.2 KRT08700.1 OUUW01000002 SPP77529.1 CH480817 EDW50811.1 CH940647 EDW68736.1 KRF83994.1 AE014296 AAF50588.4 AGB94199.1 JR038769 AEY58401.1 CH954178 EDV50502.1 CM000159 EDW93729.1 UFQT01000369 SSX23613.1 CH963847 EDW73391.2 CH902618 EDV40566.1 CVRI01000050 CRK99406.1 KQ981272 KYN43860.1 KQ982691 KYQ52210.1 KB631768 ERL85992.1 KQ980765 KYN13360.1 KQ414855 KOC60100.1 KQ976749 KYN08772.1 QOIP01000001 RLU26872.1 KQ976641 KYM78810.1 CM002912 KMY98048.1 ADTU01006076 ADTU01006077 ADTU01006078 ADTU01006079 ADTU01006080 GEZM01072759 JAV65410.1 KK852721 KDR17747.1 CP012525 ALC44409.1 AAAB01008960 EAA11102.4 CH933816 EDW17233.2 CM000363 EDX09538.1 KK107242 EZA54548.1 GL449799 EFN81951.1 KQ435841 KOX71305.1 BT011431 AAR99089.1 ATLV01022998 KE525339 KFB48238.1 EDO63740.1 KQ761906 OAD56453.1 ABLF02034728 ABLF02034739 ABLF02034740 ABLF02057270 GFXV01000680 MBW12485.1 GL762140 EFZ21915.1 AXCM01010389 ADMH02001139 ETN63965.1 AXCN02000446 APCN01000131 GGMS01007130 MBY76333.1 JXUM01091842 JXUM01091843 KQ563946 KXJ73031.1 CH477250 EAT46054.1 JXJN01019032 JXJN01019033 JXJN01019034 DS231945 EDS28323.1 CH479244 EDW37712.1 DS235149 EEB12523.1 GL732524 EFX89361.1 NEVH01007814 PNF35637.1 LBMM01001340 KMQ96482.1 NEDP02004422 OWF45641.1 PZQS01000001 PVD39324.1 KB202953 ESO87159.1 GEGO01002562 JAR92842.1 JH818302 EKC23238.1 AMQN01007080 KB299905 ELU07462.1 GGLE01003042 MBY07168.1 ABJB010047782 ABJB010056371 ABJB010060520 ABJB010093577 ABJB010181485 ABJB010222402 ABJB010363018 ABJB010448035 ABJB010514674 ABJB010582315 DS758678 EEC08524.1 HACA01004114 CDW21475.1 AHAT01029956 AHAT01029957
Proteomes
UP000218220
UP000053268
UP000053240
UP000007266
UP000019118
UP000092444
+ More
UP000092445 UP000078200 UP000001070 UP000092443 UP000037069 UP000001819 UP000268350 UP000001292 UP000008792 UP000000803 UP000008711 UP000192221 UP000002282 UP000007798 UP000007801 UP000183832 UP000095300 UP000078541 UP000075809 UP000030742 UP000078492 UP000053825 UP000078542 UP000279307 UP000078540 UP000005205 UP000091820 UP000027135 UP000092553 UP000007062 UP000009192 UP000000304 UP000053097 UP000008237 UP000053105 UP000030765 UP000007819 UP000075880 UP000075883 UP000075882 UP000075903 UP000076407 UP000069272 UP000000673 UP000075920 UP000075885 UP000075886 UP000075884 UP000075840 UP000076408 UP000069940 UP000249989 UP000075902 UP000008820 UP000075901 UP000075900 UP000092460 UP000002320 UP000008744 UP000075881 UP000009046 UP000095301 UP000000305 UP000235965 UP000085678 UP000036403 UP000242188 UP000245119 UP000030746 UP000005408 UP000014760 UP000001555 UP000018468
UP000092445 UP000078200 UP000001070 UP000092443 UP000037069 UP000001819 UP000268350 UP000001292 UP000008792 UP000000803 UP000008711 UP000192221 UP000002282 UP000007798 UP000007801 UP000183832 UP000095300 UP000078541 UP000075809 UP000030742 UP000078492 UP000053825 UP000078542 UP000279307 UP000078540 UP000005205 UP000091820 UP000027135 UP000092553 UP000007062 UP000009192 UP000000304 UP000053097 UP000008237 UP000053105 UP000030765 UP000007819 UP000075880 UP000075883 UP000075882 UP000075903 UP000076407 UP000069272 UP000000673 UP000075920 UP000075885 UP000075886 UP000075884 UP000075840 UP000076408 UP000069940 UP000249989 UP000075902 UP000008820 UP000075901 UP000075900 UP000092460 UP000002320 UP000008744 UP000075881 UP000009046 UP000095301 UP000000305 UP000235965 UP000085678 UP000036403 UP000242188 UP000245119 UP000030746 UP000005408 UP000014760 UP000001555 UP000018468
Interpro
SUPFAM
SSF56601
SSF56601
Gene 3D
ProteinModelPortal
A0A2A4K676
A0A194Q1W1
A0A194QP76
D6WGN2
N6TXN5
A0A1B0G330
+ More
A0A1A9ZF16 A0A1A9V8Y9 B4J1Z0 A0A1A9Y5I4 A0A0L0CEF3 Q29E82 A0A3B0JUZ2 B4HV40 B4LC91 Q9VS41 V9ID48 B3NFG0 A0A1W4UGT6 B4PK49 A0A336M3V5 B4MLD3 B3M4Z2 A0A1J1IK11 A0A1I8PCN0 A0A195FTC2 A0A151WWG4 U4TYT9 A0A195DKD8 A0A0L7QN91 A0A195D7D9 A0A3L8E2W9 A0A195B3G4 A0A0J9RQE5 A0A158P192 A0A1A9W7M7 A0A1Y1KZ13 A0A067RCT7 A0A0M3QWM5 Q7Q6J0 B4L958 B4QKB5 A0A026WF15 E2BQS6 A0A0M8ZUB9 Q6NN63 A0A084WDE4 A7UTW6 A0A310SJL0 J9JVL3 A0A2H8TEI2 E9IBZ6 A0A182IWZ2 A0A182MIJ4 A0A182KQ24 A0A182V040 A0A182XMG4 A0A182FDW1 W5JM45 A0A182WL80 A0A182PIG1 A0A182QI95 A0A182NJH4 A0A182I294 A0A182YJA7 A0A2S2QF62 A0A182GVW5 A0A182TJ31 Q17HF2 A0A182S617 A0A182R468 A0A1B0BRC2 B0WI97 B4HAT0 A0A182K8M0 E0VGL7 A0A1I8MMB7 E9FTH5 A0A2J7R494 A0A1S3HNM7 A0A1S3HNN6 A0A0J7L115 A0A210QA78 A0A2T7Q0X1 V4BE93 A0A147BQR0 K1PNI3 R7UU63 A0A2R5LCG0 B7PPK2 A0A0K2T6Q5 W5M7U2 W5M7S9
A0A1A9ZF16 A0A1A9V8Y9 B4J1Z0 A0A1A9Y5I4 A0A0L0CEF3 Q29E82 A0A3B0JUZ2 B4HV40 B4LC91 Q9VS41 V9ID48 B3NFG0 A0A1W4UGT6 B4PK49 A0A336M3V5 B4MLD3 B3M4Z2 A0A1J1IK11 A0A1I8PCN0 A0A195FTC2 A0A151WWG4 U4TYT9 A0A195DKD8 A0A0L7QN91 A0A195D7D9 A0A3L8E2W9 A0A195B3G4 A0A0J9RQE5 A0A158P192 A0A1A9W7M7 A0A1Y1KZ13 A0A067RCT7 A0A0M3QWM5 Q7Q6J0 B4L958 B4QKB5 A0A026WF15 E2BQS6 A0A0M8ZUB9 Q6NN63 A0A084WDE4 A7UTW6 A0A310SJL0 J9JVL3 A0A2H8TEI2 E9IBZ6 A0A182IWZ2 A0A182MIJ4 A0A182KQ24 A0A182V040 A0A182XMG4 A0A182FDW1 W5JM45 A0A182WL80 A0A182PIG1 A0A182QI95 A0A182NJH4 A0A182I294 A0A182YJA7 A0A2S2QF62 A0A182GVW5 A0A182TJ31 Q17HF2 A0A182S617 A0A182R468 A0A1B0BRC2 B0WI97 B4HAT0 A0A182K8M0 E0VGL7 A0A1I8MMB7 E9FTH5 A0A2J7R494 A0A1S3HNM7 A0A1S3HNN6 A0A0J7L115 A0A210QA78 A0A2T7Q0X1 V4BE93 A0A147BQR0 K1PNI3 R7UU63 A0A2R5LCG0 B7PPK2 A0A0K2T6Q5 W5M7U2 W5M7S9
PDB
3PFQ
E-value=1.23573e-13,
Score=190
Ontologies
GO
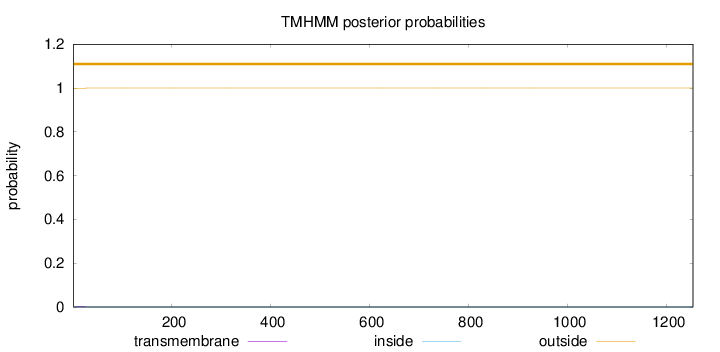
Topology
Length:
1253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04254
Exp number, first 60 AAs:
0.04169
Total prob of N-in:
0.00209
outside
1 - 1253
Population Genetic Test Statistics
Pi
246.957178
Theta
17.875249
Tajima's D
0.839092
CLR
0.555553
CSRT
0.620018999050048
Interpretation
Uncertain
