Gene
KWMTBOMO06647
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.802 Nuclear Reliability : 1.262
Sequence
CDS
ATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTCATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTCTCTTCTTCGACGTCGCAAAAGCGTTCGACAAAGTCTGGCACAATGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTCAACGATATTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCTAGTAGAAATAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTATTTCAGAGGGGAAGCTCCACACGGATTTCCTCCCGGATTAGGAGGAGGAATCTCACACCCCCGATTACTCTCTTTAGACAACCCATACCCTGGGCCAGGAAGGTCAAGTACCTGGGCGTTACCCTGGATGCATCGATGACATTCCGCCCGCATATAAAATCAGTCCGTGACCGTGCCGCGTTTATTCTCGGTAGACTCTACCCCATGATCTGTAAGCGGAGTAAAATGTCCCTTCGGAACAAGGTGACACTTTACAAAACTTGCATAAGGCCCGTCATGACTTACGCGAGTGTGGTGTTCGCTCACGCGGCCCGCACACACATAGACACCCTCCAATCCCTACAATCCCGCTTTTGCAGGTTAGCTGTCGGGGCTCCGTGGTTCGTGAGGAACGTTGACCTACACGACGACCTGGGCCTCGAATCAATTCGGAAATACATGAAGTCAGCGTCGGAACGATACTTCGATAAGGCTATGCGTCATGATAATCGCCTTATCGTTGCCGCCGCTGACTACTCCCCGAATCCTGATCATGCAGGAGCCAGTCACCGTCGACGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACCTTTGCATTAGATGCCTTCAGCTCTAATACTAGGGGCAGGCTTAGGGACCCCGTTAACGCCCGTCGACGGACCGTCGAGGCGATCCGCCCGGTCCTCTCGGACTGGGTGAATCGCGACCGAGGACGTCTCACCTTCCGAGCGACGCAGGTGCTCACGGGACACGGCTGTTTCGGTCGCTACCTGCACCTCGTCGCCCGGAGGGAGCCGACGCCGAAGTGCCACCACTGCAGTGGCTGCAACGAGGACACGGCGGAGCACACGCTCGCGTACTGCCCCGCTTTCGCGGAGCAGCGCCGCGTCCTCGTTGCAAAAATAGGACCGGACTTGTCGCTTCCGACCGTCGTGGCTACGATACTCGGCAGCGACGAGTCCTGGCAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGCAGAAGGAGGCGGCGGAACGGGAGAGGGAGAGCTCTTCTTCCCTCTCGGCGCCGTGCCGCCGCCGTCGGGCCGGGGGCCGGAGGAGGGCGTTTGTCCAGCTCCAGCCCCTATGA
Protein
MLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGALFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDIPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQRGSSTRISSRIRRRNLTPPITLFRQPIPWARKVKYLGVTLDASMTFRPHIKSVRDRAAFILGRLYPMICKRSKMSLRNKVTLYKTCIRPVMTYASVVFAHAARTHIDTLQSLQSRFCRLAVGAPWFVRNVDLHDDLGLESIRKYMKSASERYFDKAMRHDNRLIVAAADYSPNPDHAGASHRRRPRHVLTDPSDPITFALDAFSSNTRGRLRDPVNARRRTVEAIRPVLSDWVNRDRGRLTFRATQVLTGHGCFGRYLHLVARREPTPKCHHCSGCNEDTAEHTLAYCPAFAEQRRVLVAKIGPDLSLPTVVATILGSDESWQAMLDFCESTISQKEAAERERESSSSLSAPCRRRRAGGRRRAFVQLQPL
Summary
Uniprot
Q93137
Q9XXW0
Q9BPQ0
A0A0N0PA09
A0A0N1PFV1
O96546
+ More
A0A1Y1NA23 V5GNF3 V5GNJ2 A0A139WA11 A0A2S2PH29 X1WL89 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RIW7 A0A2J7Q4F7 A0A2J7R4B7 A0A2J7Q059 A0A2J7RM10 A0A2J7PMP6 A0A2J7QUJ9 A0A2S2RAN7 A0A2J7RB54 A0A2J7RPU2 A0A2J7PMP9 A0A2J7RS77 A0A023F742 X1WZH0 A0A2H8TWD9 A0A2J7QEZ4 A0A2J7Q5G9 A0A2J7R2E1 A0A2J7QFL0 A0A2J7Q0Q6 A0A2J7R6U1 A0A2P8Z520 A0A2J7RKP3 A0A2J7QR37 A0A2J7Q6J2
A0A1Y1NA23 V5GNF3 V5GNJ2 A0A139WA11 A0A2S2PH29 X1WL89 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RIW7 A0A2J7Q4F7 A0A2J7R4B7 A0A2J7Q059 A0A2J7RM10 A0A2J7PMP6 A0A2J7QUJ9 A0A2S2RAN7 A0A2J7RB54 A0A2J7RPU2 A0A2J7PMP9 A0A2J7RS77 A0A023F742 X1WZH0 A0A2H8TWD9 A0A2J7QEZ4 A0A2J7Q5G9 A0A2J7R2E1 A0A2J7QFL0 A0A2J7Q0Q6 A0A2J7R6U1 A0A2P8Z520 A0A2J7RKP3 A0A2J7QR37 A0A2J7Q6J2
EMBL
U07847
AAA17752.1
AB018558
BAA76304.1
AB055391
BAB21761.1
+ More
KQ459259 KPJ02068.1 KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GALX01005359 JAB63107.1 GALX01005299 JAB63167.1 KQ971729 KYB24751.1 GGMR01016114 MBY28733.1 ABLF02019349 ABLF02041900 NEVH01006736 PNF36716.1 GEZM01053022 JAV74219.1 NEVH01003017 PNF40781.1 NEVH01018383 PNF23463.1 NEVH01007443 PNF35684.1 NEVH01020071 PNF21969.1 NEVH01021586 NEVH01006600 NEVH01002584 PNF19589.1 PNF37419.1 PNF41868.1 NEVH01023962 PNF17612.1 NEVH01010546 PNF32261.1 GGMS01017914 MBY87117.1 NEVH01005938 PNF38077.1 NEVH01001355 PNF42855.1 NEVH01023961 PNF17613.1 NEVH01000280 PNF43672.1 GBBI01001693 JAC17019.1 ABLF02034467 GFXV01006424 MBW18229.1 NEVH01015305 NEVH01006671 PNF27154.1 PNF37357.1 NEVH01017559 PNF23833.1 NEVH01007838 PNF34986.1 NEVH01014858 PNF27387.1 NEVH01019964 PNF22161.1 NEVH01006754 PNF36554.1 PYGN01000192 PSN51582.1 NEVH01002716 PNF41415.1 NEVH01011985 PNF31054.1 NEVH01017470 PNF24199.1
KQ459259 KPJ02068.1 KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GALX01005359 JAB63107.1 GALX01005299 JAB63167.1 KQ971729 KYB24751.1 GGMR01016114 MBY28733.1 ABLF02019349 ABLF02041900 NEVH01006736 PNF36716.1 GEZM01053022 JAV74219.1 NEVH01003017 PNF40781.1 NEVH01018383 PNF23463.1 NEVH01007443 PNF35684.1 NEVH01020071 PNF21969.1 NEVH01021586 NEVH01006600 NEVH01002584 PNF19589.1 PNF37419.1 PNF41868.1 NEVH01023962 PNF17612.1 NEVH01010546 PNF32261.1 GGMS01017914 MBY87117.1 NEVH01005938 PNF38077.1 NEVH01001355 PNF42855.1 NEVH01023961 PNF17613.1 NEVH01000280 PNF43672.1 GBBI01001693 JAC17019.1 ABLF02034467 GFXV01006424 MBW18229.1 NEVH01015305 NEVH01006671 PNF27154.1 PNF37357.1 NEVH01017559 PNF23833.1 NEVH01007838 PNF34986.1 NEVH01014858 PNF27387.1 NEVH01019964 PNF22161.1 NEVH01006754 PNF36554.1 PYGN01000192 PSN51582.1 NEVH01002716 PNF41415.1 NEVH01011985 PNF31054.1 NEVH01017470 PNF24199.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Q93137
Q9XXW0
Q9BPQ0
A0A0N0PA09
A0A0N1PFV1
O96546
+ More
A0A1Y1NA23 V5GNF3 V5GNJ2 A0A139WA11 A0A2S2PH29 X1WL89 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RIW7 A0A2J7Q4F7 A0A2J7R4B7 A0A2J7Q059 A0A2J7RM10 A0A2J7PMP6 A0A2J7QUJ9 A0A2S2RAN7 A0A2J7RB54 A0A2J7RPU2 A0A2J7PMP9 A0A2J7RS77 A0A023F742 X1WZH0 A0A2H8TWD9 A0A2J7QEZ4 A0A2J7Q5G9 A0A2J7R2E1 A0A2J7QFL0 A0A2J7Q0Q6 A0A2J7R6U1 A0A2P8Z520 A0A2J7RKP3 A0A2J7QR37 A0A2J7Q6J2
A0A1Y1NA23 V5GNF3 V5GNJ2 A0A139WA11 A0A2S2PH29 X1WL89 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RIW7 A0A2J7Q4F7 A0A2J7R4B7 A0A2J7Q059 A0A2J7RM10 A0A2J7PMP6 A0A2J7QUJ9 A0A2S2RAN7 A0A2J7RB54 A0A2J7RPU2 A0A2J7PMP9 A0A2J7RS77 A0A023F742 X1WZH0 A0A2H8TWD9 A0A2J7QEZ4 A0A2J7Q5G9 A0A2J7R2E1 A0A2J7QFL0 A0A2J7Q0Q6 A0A2J7R6U1 A0A2P8Z520 A0A2J7RKP3 A0A2J7QR37 A0A2J7Q6J2
Ontologies
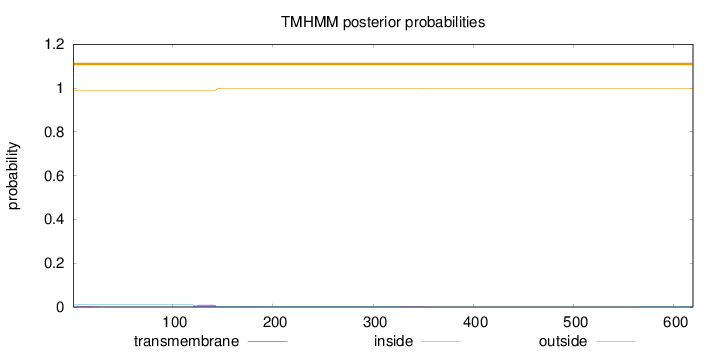
Topology
Length:
619
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28271
Exp number, first 60 AAs:
0.02884
Total prob of N-in:
0.01118
outside
1 - 619
Population Genetic Test Statistics
Pi
19.96156
Theta
18.614759
Tajima's D
-1.945731
CLR
172.163012
CSRT
0.0145492725363732
Interpretation
Uncertain
