Pre Gene Modal
BGIBMGA011702
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.82
Sequence
CDS
ATGGAAAAAGAACACCAGCCAGACTCGATGGCTACAATAACTATGAAGCCGGAATATCCGCCTTCTGAAGTATACAGCACATCAGAACCGCCACCGGCTTATCGGCACAGGGTGTCAACTTCGGTCCAGATCGCGAAGATTGCAGCACTAACAGTGGTCGCTTCCTCCTTCATCTTGGGAACCTTTATATTGGCTTCGAGTTGGGTAGCAGCTCGTTCATCATGCCATCAGCTAGAGCAGCTGGATGCAATGCTCGACAAAGAACTCGCTTTGGAAGGCAGAGCCTACGGAAATGATGCCCTGGTAGCGGATGAGCCTTTGCCACTAGCAAATGCCCACGCTTTGCATGGAGTACCGCCCATGCTGTCTTCAGTGTTACCTGAGACGAGCCAGCCGTCATCTTCACGACCCAGTCTCTTCAAAGACGACGCACTCAACCATGCCGAGTCCAAAATAAACGAAGACAAATTGCAGAAAATTGACGACGATAAGAATGACTCACCAAACTCGAGTGATGAGAGTCCTGAGTCCGACAGTTCTGCTGAAGAGGACGACGAACTTGAAGCGATTAGACCGATGTTCAAATTGCCTATTCAGTTTGACCTTGATGAGTTGGCGGGCGCTTTCTTAGCAAACAACCAAAAAGGCCGTATGAACTGCGTGGTGGAGCGTCGTAACGACGACCCCATGGAGAGACGTTTTCCCTTCAACATTCTAGGCGCGTTCGCTCCCCGCAGCGCTCATGCCGAACGCGTAGCCATCATCTGCCACGGAGGAGATGAGCCCAGGGCAATCAATTTCCCGGAGCCACAGTCCCAGGTATTTGCCTTCCCGTTGACCGGTCCAGTACGCCTCCCTCTGCGTCCTCAGCCAGAGACACCTTCCGGTCCTGAACCCAGGATGCCTTTTGGACCTGGTCCTGACGGTAGAATGCCACCTTTCGCACAAGCTCCTGAACAAAGAATGCCGCCCTTCGCACAAGCTCCTGAACAAAGAATGCCGCCCTTCGCTCCACTAGGACCGATGGCACCAGTTAGCCGCCAGGCGTCCTTCCCGCTGCCGAGATTCACGCTGCGAACAGACTTAATTCCCGGCAGCTTTACTATCCCTGGCTACGGTAGCAGTCATGGTAACGGTAAGGCAGGGGGTGCGAAGAATCCCCGGGCTATGGACAGCTACCACCAAATGACCCTGGCGACTCTCAACACCCGGACCCTGCGGCTTGACGAACATATTGAGGAACTGGAAGTGGAATTAGGAAGGATACGATGGCACGTACTAGGGTTGTGCGAAGTCCGAAGACAGGGGGAGGACGTCATAACTCTGGACTCAGGCCACCTATTCTACTTCCGGGAAGGAGACCAACCTGGACAAGGTGGAGTTGGCTTTCTCGTCAATAAAATCCTCACAGACAATATTGTAGAGGTATCCAGTGTGTCGACACGGGTAGCGTACCTCGTAATTAAGCTCACCGAAAGGTACTCCCTTAAGGTGGTGCAAGTATACGCACCGACTTCGTCATACTCGGATGACGATGTGGAGGTTGTGTACGACGATATTACCAGGGCTCTGCACGATACCACAAAAGCCCACTTCAATGTTGTCATGGGAGATTTTAACGCTAAAGTGGGAGTACAGACTTGCGACGAATCGGTGCTAGGATCCTTCGGTTATGGATGTAGGAACCACAGGGGGCAAATGCTAGTCAACTTCCTTCAGCGCGAGAGGCTCTTTCTAATGAACTCGTTCTTTAAAAAGAAGCCACAAAGGAAGTGGACTTGGCTAAGCCCCGACACTGTGACGAAAAACGAGATAGACTTTATTATGACGGACAAGAAGCATATATTCAGGGATGTCTCTGTGATCAGCAGGTTCAATACCGGTAGTGATCACCGACTTGTCAGAGGCACACTGAATATGGACTTCAAGTTAGAACGCACTCGTCTAATAAAGTCTACGCTTCGACCCAGCCTACATCACATGGCTGTAGATCCGGAGCAATTCCAGCTAGATCTCCACAACCGATTCGCTGCCTTGGAAACCACCACAGACGTAGACGAGGCCTTAGATGGCGTGGTGAGGACACTGAGACAGGAGGGACAGAGACTGTGTCCAAAGGGTCGTACAGGCAGAACATCAAAGTTATCATCTGAGACTCTCCGGCTAATGGAAGATAGGCGAGAAAATACACAGGCCACACAATCAGAGAAGGTGGCTCTTAATAAGAAAATCAGGACGCTGATACGACGCGATCTCCGCCGGTACAATACTCAGCGGATCAATGACGCAATCGAGCAAAACCGAGGAGCCAAGGTGTTCGTGCAGCGTCTTGGTAAAAGCCACCCTACAAAACTGACCAAAAGGGATGGCCAAATTCTCTCCTCCAAACCGGAGATTCTCAAGGAAATTGAGGACTTCTACGGAGGCCTCTATACTTCCAAAGCAACTAAGCCTGTCTCCGACCCAGCAGATGTCCGAGCCACACTGTCCCGCCACTACACCGACGAGCTGCCACAGATTAGTGTAGACGAGATCGGCATGGCTCTTGAAAATCTTAAAAACGGAAAGGCCCCGGGGGAGGATGGTGTTACTTCTGAGCTCTTGAAAGCAGGAGGTCTTCCTGTTATTAGGGAGCTCCAGAAGCTCTTCAACACTGTCCTCTTTACCGGTAAAACTCCAAAGGCGTGGAGCAGGAGCATAGTTGTCCTTTTCTTTAAGAAAGGAGATAAGACACTTTTAAAGAATTATCGCCCCATCTCGCTCCTGAGCCACGTCTACAAATTGTTTTCAAGAGTCATAACAAATCGTCTTGCTCAGAGACTTGACGACTTCCAGCCTCCGGAACAGGCAGGATTCCGAAGAGGATTTGGTACCGTGGACCACATTCACACAGTTCGGCAGATTATACAGAAGACTGAAGAGTATAATCTGCCACTATGTCTGGCGTTTGTGGACTACGAGAAAGCCTTTGACTCGATCGAGACCTGGGCTGTTCTGGAATCTTTGCAGCGGTGTCAAGCTGACTGGCGATACATCGATGCACTAAGATGCCTGTACGATGCCGCTACAATGACCGTCCAAGTACAGAAAGACCAGACGAGACCAATCCAGTTGCGCAGAGGTGTAAGACAGGGGGATATTATATCCCCGAAACTGTTTACAAACGCACTGGAGGATGTCTTCAAGACTCTGGACTGGAACGGACGCGGCATCAACATAAACGGCGAGTACATCTCACATCTTCGCTTTGCCGATGATATCGTCATCATGGCGGAATCGCTGCAGGACCTACAAGAAATGGTGCACAGCCTTAATACTGCATCCCAACGGGTTGGCCTCGGAATGAACTTGGATAAAACCAAGGTAATGTTTAACGGAAACGTCATTCCGAGACCAATCGATGTTGGCGGTACACCTCTCGGAGTTGTTCAAGAGTATGTCTACCTGGGACAGACCCTACAACTAGGTAGAAACAACTTTGAGAAGGAAGTGACCAGAAGGATTCAGTTGGGCTGGGCAGCGTTCGGGAAGCTTCGTCAGGTCTTTTCGTCGCCCATACCACAATGTCTGAAGACAAAGGTCTATAATCAGTGCGTCCTACCCGTGCTTACCTACGGAGCCGAAACGTGGACACTGACTGTGCGACTGGTCCACCAATTAAAAGTTACTCAGCGAGCTATGGAGCGAGCTATGCTCGGTTTGTCATTGCGAGACCGAATACGCAATGAGGTCATTCGGGCAAAAACCAAAGTGATCGATATAGCTCGCAGAGTTAGTAAGCTGAAGTGGCAGTGGGCTGGTCACATATGTCGCAGAACCGATAACCGTTGGGGTAGACGTGTTCTGGAGTGGAGACCGCGAACCGGCAAACGTAGTGTAGGACGCCCTCCTGCTAGATGGAGCGACGACCTGCGCAAGGCAGCTGGCAAAGAGAGCCACAACAAACAAAATGATAAGGTGTACGCACACAGTATTGAAGAAGCGAGCAACCGTATTCCGCGTGTCCAACGAGGTCATTTTCCATCCTCGCTCATGGTATGGTTGGGAGTTTCTTATTGGGGCTTAACAGAGGTACATTTTTGTAAGAAAGGTGTAAAAACGAATGCAGTTGTGTATCAAAATACAGTCCTGACGAACCTTGTGGAACCTGTTTCTCATACCATGTTCAATAACAGGCACTGGGTATTCCAACAAGATTGGGCGCCAGCTCATAGAGCGAAGAGCACACCAGACTGGCTGGCGGCGTGTGAAATCGACTTCATCCGGCACGAAGACTGGCCCTCTTCCAGTCCAGATTTGAATCCGTTAGATTACAAGATATGGCAACACTTGGAGGAAAAGGCGTGCTCAAAGCCTCATCCCAATTTGGAGTCACTCAAGACATCCTTGATTAAGGCAGCCGCCGATATTGACATGGACCTCGTTCGTGCTGCGATAGACGACTGGCCGCGCAGATTGAAGGCCTGTATTCAAAATCACGGAGGTCATTTTGAATAA
Protein
MEKEHQPDSMATITMKPEYPPSEVYSTSEPPPAYRHRVSTSVQIAKIAALTVVASSFILGTFILASSWVAARSSCHQLEQLDAMLDKELALEGRAYGNDALVADEPLPLANAHALHGVPPMLSSVLPETSQPSSSRPSLFKDDALNHAESKINEDKLQKIDDDKNDSPNSSDESPESDSSAEEDDELEAIRPMFKLPIQFDLDELAGAFLANNQKGRMNCVVERRNDDPMERRFPFNILGAFAPRSAHAERVAIICHGGDEPRAINFPEPQSQVFAFPLTGPVRLPLRPQPETPSGPEPRMPFGPGPDGRMPPFAQAPEQRMPPFAQAPEQRMPPFAPLGPMAPVSRQASFPLPRFTLRTDLIPGSFTIPGYGSSHGNGKAGGAKNPRAMDSYHQMTLATLNTRTLRLDEHIEELEVELGRIRWHVLGLCEVRRQGEDVITLDSGHLFYFREGDQPGQGGVGFLVNKILTDNIVEVSSVSTRVAYLVIKLTERYSLKVVQVYAPTSSYSDDDVEVVYDDITRALHDTTKAHFNVVMGDFNAKVGVQTCDESVLGSFGYGCRNHRGQMLVNFLQRERLFLMNSFFKKKPQRKWTWLSPDTVTKNEIDFIMTDKKHIFRDVSVISRFNTGSDHRLVRGTLNMDFKLERTRLIKSTLRPSLHHMAVDPEQFQLDLHNRFAALETTTDVDEALDGVVRTLRQEGQRLCPKGRTGRTSKLSSETLRLMEDRRENTQATQSEKVALNKKIRTLIRRDLRRYNTQRINDAIEQNRGAKVFVQRLGKSHPTKLTKRDGQILSSKPEILKEIEDFYGGLYTSKATKPVSDPADVRATLSRHYTDELPQISVDEIGMALENLKNGKAPGEDGVTSELLKAGGLPVIRELQKLFNTVLFTGKTPKAWSRSIVVLFFKKGDKTLLKNYRPISLLSHVYKLFSRVITNRLAQRLDDFQPPEQAGFRRGFGTVDHIHTVRQIIQKTEEYNLPLCLAFVDYEKAFDSIETWAVLESLQRCQADWRYIDALRCLYDAATMTVQVQKDQTRPIQLRRGVRQGDIISPKLFTNALEDVFKTLDWNGRGININGEYISHLRFADDIVIMAESLQDLQEMVHSLNTASQRVGLGMNLDKTKVMFNGNVIPRPIDVGGTPLGVVQEYVYLGQTLQLGRNNFEKEVTRRIQLGWAAFGKLRQVFSSPIPQCLKTKVYNQCVLPVLTYGAETWTLTVRLVHQLKVTQRAMERAMLGLSLRDRIRNEVIRAKTKVIDIARRVSKLKWQWAGHICRRTDNRWGRRVLEWRPRTGKRSVGRPPARWSDDLRKAAGKESHNKQNDKVYAHSIEEASNRIPRVQRGHFPSSLMVWLGVSYWGLTEVHFCKKGVKTNAVVYQNTVLTNLVEPVSHTMFNNRHWVFQQDWAPAHRAKSTPDWLAACEIDFIRHEDWPSSSPDLNPLDYKIWQHLEEKACSKPHPNLESLKTSLIKAAADIDMDLVRAAIDDWPRRLKACIQNHGGHFE
Summary
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
PDB
5CR4
E-value=4.26962e-09,
Score=152
Ontologies
KEGG
GO
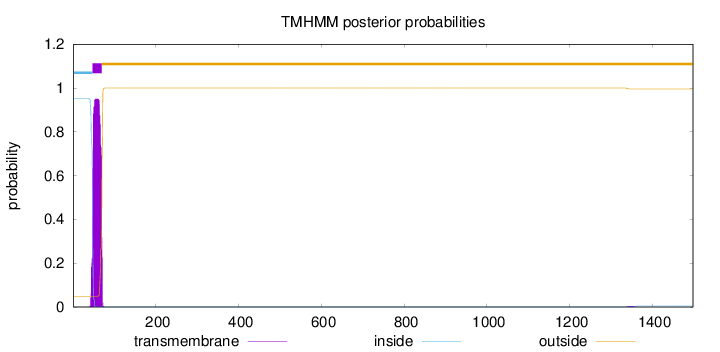
Topology
Length:
1498
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.63481
Exp number, first 60 AAs:
13.33609
Total prob of N-in:
0.95101
POSSIBLE N-term signal
sequence
inside
1 - 46
TMhelix
47 - 69
outside
70 - 1498
Population Genetic Test Statistics
Pi
163.046223
Theta
157.789095
Tajima's D
-0.543671
CLR
157.298942
CSRT
0.231738413079346
Interpretation
Possibly Positive selection
