Gene
KWMTBOMO06643 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011779
Annotation
PREDICTED:_probable_prefoldin_subunit_4_[Plutella_xylostella]
Full name
Prefoldin subunit 4
Location in the cell
Cytoplasmic Reliability : 1.98 Nuclear Reliability : 2.381
Sequence
CDS
ATGTCAACTACAGCTAAAGGAACTTTTCAGCCAGACTCTGATGTACACATTTCATATGAAGATCAACAAAAAATTAACAAGTTTGCTCGTCTCAATGCTAAAGTTGACGATTATAAGGATGAGCTCAAAGTAAAACAAAACGATGTAAAGAACCTAGAAGAAGCAGTTGAGGAACTGAGCCTTGCTGATGATTCTGAAAAAATCCCTTACCTTATTGGTGAAATTTTTATTTGTCAGAATTTGGAAATTACTTTGAAAAATCTTGAGGAAGCTAAAGCAAAAAAAATACATGAAATTCATGATTTAGAAGAAAAATGTGAGGAACTAAAATCACAAATGAGTGACTTGAAAGCACACTTGTATGGAAAATTTGGTAGCCACATTAACTTGGAAAATGAGGAAGATTGA
Protein
MSTTAKGTFQPDSDVHISYEDQQKINKFARLNAKVDDYKDELKVKQNDVKNLEEAVEELSLADDSEKIPYLIGEIFICQNLEITLKNLEEAKAKKIHEIHDLEEKCEELKSQMSDLKAHLYGKFGSHINLENEED
Summary
Description
Binds specifically to cytosolic chaperonin (c-CPN) and transfers target proteins to it. Binds to nascent polypeptide chain and promotes folding in an environment in which there are many competing pathways for nonnative proteins.
Subunit
Heterohexamer of two PFD-alpha type and four PFD-beta type subunits.
Similarity
Belongs to the prefoldin subunit beta family.
Uniprot
H9JQH3
A0A2A4JBT4
A0A2H1VXB2
S4PMN4
A0A3S2NW85
A0A212ELI2
+ More
A0A194QQC4 I4DLD8 A0A0L7L117 D6X221 A0A2J7Q722 V5I7Y6 U4UKA1 A0A067R1Q5 N6SV95 J3JXB8 E2BHI5 A0A195CTQ6 R4UKZ0 A0A1B6MDB0 A0A232EYK4 K7JAQ8 A0A2S2NGK1 C4WSI6 A0A0V0G5R6 A0A224XZW6 A0A0T6B6I4 A0A1L8DU37 A0A0P4VK23 U5EZA6 T1DHD4 A0A2M3Z6Z5 A0A1B6HCG7 A0A182WWI6 A0A182UPF3 A0A182JRP3 A0A182I6R7 A0A0N0U709 A0A182MCN7 A0A182WBI9 A0A182SL99 A0A182UDD8 A0A1Q3FBZ0 A0A182Y2C2 Q7QIM0 A0A182PII6 A0A1B6G7J4 R4WT38 W5JV23 A0A182FEL8 A0A2M4C2Z2 A0A0L7QQ59 A0A1B0D5V5 A0A1W4WHZ9 A0A2M4AGV2 A0A182RJL6 A0A195BP16 A0A195FR15 A0A158NTS0 A0A151WX69 A0A0J7NNA4 A0A182QFC1 A0A026W939 F4WJ11 A0A182HA99 Q17IJ1 A0A151J594 B0W5L5 A0A151JNR5 A0A0K8TP25 A0A182N9A1 A0A1Y1KJW0 A0A3L8DA49 A0A1S3DCZ7 A0A2A3EG24 V9ILX5 A0A088ATX6 A0A1B0GII4 A0A182JEN9 A0A336N5J2 A0A1B6DQ76 A0A084W148 T1GKH0 T1J251 B4PIT1 A0A336L8G3 T1HUH4 B3NGB1 A0A0L0CCD3 W8B6K5 B4MM11 A0A3B0JW25 Q2LZL9 B4H3B9 A0A0K8UTS8 A0A034VEH7 B4IX17 A0A1A9UP26 B3M3Q7 B4KXE6
A0A194QQC4 I4DLD8 A0A0L7L117 D6X221 A0A2J7Q722 V5I7Y6 U4UKA1 A0A067R1Q5 N6SV95 J3JXB8 E2BHI5 A0A195CTQ6 R4UKZ0 A0A1B6MDB0 A0A232EYK4 K7JAQ8 A0A2S2NGK1 C4WSI6 A0A0V0G5R6 A0A224XZW6 A0A0T6B6I4 A0A1L8DU37 A0A0P4VK23 U5EZA6 T1DHD4 A0A2M3Z6Z5 A0A1B6HCG7 A0A182WWI6 A0A182UPF3 A0A182JRP3 A0A182I6R7 A0A0N0U709 A0A182MCN7 A0A182WBI9 A0A182SL99 A0A182UDD8 A0A1Q3FBZ0 A0A182Y2C2 Q7QIM0 A0A182PII6 A0A1B6G7J4 R4WT38 W5JV23 A0A182FEL8 A0A2M4C2Z2 A0A0L7QQ59 A0A1B0D5V5 A0A1W4WHZ9 A0A2M4AGV2 A0A182RJL6 A0A195BP16 A0A195FR15 A0A158NTS0 A0A151WX69 A0A0J7NNA4 A0A182QFC1 A0A026W939 F4WJ11 A0A182HA99 Q17IJ1 A0A151J594 B0W5L5 A0A151JNR5 A0A0K8TP25 A0A182N9A1 A0A1Y1KJW0 A0A3L8DA49 A0A1S3DCZ7 A0A2A3EG24 V9ILX5 A0A088ATX6 A0A1B0GII4 A0A182JEN9 A0A336N5J2 A0A1B6DQ76 A0A084W148 T1GKH0 T1J251 B4PIT1 A0A336L8G3 T1HUH4 B3NGB1 A0A0L0CCD3 W8B6K5 B4MM11 A0A3B0JW25 Q2LZL9 B4H3B9 A0A0K8UTS8 A0A034VEH7 B4IX17 A0A1A9UP26 B3M3Q7 B4KXE6
Pubmed
19121390
23622113
22118469
26354079
22651552
26227816
+ More
18362917 19820115 23537049 24845553 22516182 20798317 28648823 20075255 27129103 25244985 12364791 14747013 17210077 23691247 20920257 23761445 21347285 24508170 21719571 26483478 17510324 26369729 28004739 30249741 24438588 17994087 17550304 26108605 24495485 15632085 25348373
18362917 19820115 23537049 24845553 22516182 20798317 28648823 20075255 27129103 25244985 12364791 14747013 17210077 23691247 20920257 23761445 21347285 24508170 21719571 26483478 17510324 26369729 28004739 30249741 24438588 17994087 17550304 26108605 24495485 15632085 25348373
EMBL
BABH01011997
NWSH01002234
PCG68863.1
ODYU01005003
SOQ45448.1
GAIX01003465
+ More
JAA89095.1 RSAL01000137 RVE46195.1 AGBW02014056 AGBW02013014 OWR42345.1 OWR44092.1 KQ461190 KPJ07160.1 AK402106 KQ459590 BAM18728.1 KPI97385.1 JTDY01003664 KOB69188.1 KQ971371 EFA09930.1 NEVH01017447 PNF24379.1 GALX01005620 JAB62846.1 KB632254 ERL90616.1 KK852777 KDR16741.1 APGK01055197 KB741261 ENN71604.1 BT127887 AEE62849.1 GL448301 EFN84838.1 KQ977279 KYN04050.1 KC740980 AGM32804.1 GEBQ01006060 JAT33917.1 NNAY01001609 OXU23441.1 AAZX01016257 GGMR01003666 MBY16285.1 ABLF02021773 AK340205 BAH70856.1 GECL01002787 JAP03337.1 GFTR01002236 JAW14190.1 LJIG01009497 KRT82962.1 GFDF01004131 JAV09953.1 GDKW01001959 JAI54636.1 GANO01001139 JAB58732.1 GAMD01002416 JAA99174.1 GGFM01003524 MBW24275.1 GECU01035375 JAS72331.1 APCN01003693 KQ435724 KOX78382.1 AXCM01000934 GFDL01009975 JAV25070.1 AAAB01008807 EAA04281.3 GECZ01011369 JAS58400.1 AK417892 BAN21107.1 ADMH02000413 ETN66629.1 GGFJ01010523 MBW59664.1 KQ414806 KOC60626.1 AJVK01000400 GGFK01006621 MBW39942.1 KQ976435 KYM87097.1 KQ981382 KYN42349.1 ADTU01026117 KQ982673 KYQ52438.1 LBMM01003082 KMQ93990.1 AXCN02000558 KK107348 EZA52151.1 GL888181 EGI65781.1 JXUM01122301 KQ566589 KXJ69990.1 CH477239 EAT46478.1 KQ980025 KYN18204.1 DS231843 EDS35498.1 KQ978878 KYN27858.1 GDAI01001476 JAI16127.1 GEZM01081333 JAV61699.1 QOIP01000010 RLU17375.1 KZ288255 PBC30670.1 JR051397 AEY61491.1 AJWK01014150 UFQT01002611 SSX33768.1 GEDC01009478 JAS27820.1 ATLV01019208 KE525264 KFB43942.1 CAQQ02394204 JH431796 CM000159 EDW93501.1 UFQS01001300 UFQT01001300 SSX10175.1 SSX29896.1 ACPB03012087 CH954178 EDV51012.1 JRES01000611 KNC29891.1 GAMC01013897 JAB92658.1 CH963847 EDW73020.1 OUUW01000002 SPP76921.1 CH379069 EAL29488.1 CH479206 EDW30859.1 GDHF01022391 JAI29923.1 GAKP01017271 GAKP01017270 JAC41681.1 CH916366 EDV96323.1 CH902618 EDV40350.1 CH933809 EDW18632.1
JAA89095.1 RSAL01000137 RVE46195.1 AGBW02014056 AGBW02013014 OWR42345.1 OWR44092.1 KQ461190 KPJ07160.1 AK402106 KQ459590 BAM18728.1 KPI97385.1 JTDY01003664 KOB69188.1 KQ971371 EFA09930.1 NEVH01017447 PNF24379.1 GALX01005620 JAB62846.1 KB632254 ERL90616.1 KK852777 KDR16741.1 APGK01055197 KB741261 ENN71604.1 BT127887 AEE62849.1 GL448301 EFN84838.1 KQ977279 KYN04050.1 KC740980 AGM32804.1 GEBQ01006060 JAT33917.1 NNAY01001609 OXU23441.1 AAZX01016257 GGMR01003666 MBY16285.1 ABLF02021773 AK340205 BAH70856.1 GECL01002787 JAP03337.1 GFTR01002236 JAW14190.1 LJIG01009497 KRT82962.1 GFDF01004131 JAV09953.1 GDKW01001959 JAI54636.1 GANO01001139 JAB58732.1 GAMD01002416 JAA99174.1 GGFM01003524 MBW24275.1 GECU01035375 JAS72331.1 APCN01003693 KQ435724 KOX78382.1 AXCM01000934 GFDL01009975 JAV25070.1 AAAB01008807 EAA04281.3 GECZ01011369 JAS58400.1 AK417892 BAN21107.1 ADMH02000413 ETN66629.1 GGFJ01010523 MBW59664.1 KQ414806 KOC60626.1 AJVK01000400 GGFK01006621 MBW39942.1 KQ976435 KYM87097.1 KQ981382 KYN42349.1 ADTU01026117 KQ982673 KYQ52438.1 LBMM01003082 KMQ93990.1 AXCN02000558 KK107348 EZA52151.1 GL888181 EGI65781.1 JXUM01122301 KQ566589 KXJ69990.1 CH477239 EAT46478.1 KQ980025 KYN18204.1 DS231843 EDS35498.1 KQ978878 KYN27858.1 GDAI01001476 JAI16127.1 GEZM01081333 JAV61699.1 QOIP01000010 RLU17375.1 KZ288255 PBC30670.1 JR051397 AEY61491.1 AJWK01014150 UFQT01002611 SSX33768.1 GEDC01009478 JAS27820.1 ATLV01019208 KE525264 KFB43942.1 CAQQ02394204 JH431796 CM000159 EDW93501.1 UFQS01001300 UFQT01001300 SSX10175.1 SSX29896.1 ACPB03012087 CH954178 EDV51012.1 JRES01000611 KNC29891.1 GAMC01013897 JAB92658.1 CH963847 EDW73020.1 OUUW01000002 SPP76921.1 CH379069 EAL29488.1 CH479206 EDW30859.1 GDHF01022391 JAI29923.1 GAKP01017271 GAKP01017270 JAC41681.1 CH916366 EDV96323.1 CH902618 EDV40350.1 CH933809 EDW18632.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000037510 UP000007266 UP000235965 UP000030742 UP000027135 UP000019118 UP000008237 UP000078542 UP000215335 UP000002358 UP000007819 UP000076407 UP000075903 UP000075881 UP000075840 UP000053105 UP000075883 UP000075920 UP000075901 UP000075902 UP000076408 UP000007062 UP000075885 UP000000673 UP000069272 UP000053825 UP000092462 UP000192223 UP000075900 UP000078540 UP000078541 UP000005205 UP000075809 UP000036403 UP000075886 UP000053097 UP000007755 UP000069940 UP000249989 UP000008820 UP000078492 UP000002320 UP000075884 UP000279307 UP000079169 UP000242457 UP000005203 UP000092461 UP000075880 UP000030765 UP000015102 UP000002282 UP000015103 UP000008711 UP000037069 UP000007798 UP000268350 UP000001819 UP000008744 UP000001070 UP000078200 UP000007801 UP000009192
UP000037510 UP000007266 UP000235965 UP000030742 UP000027135 UP000019118 UP000008237 UP000078542 UP000215335 UP000002358 UP000007819 UP000076407 UP000075903 UP000075881 UP000075840 UP000053105 UP000075883 UP000075920 UP000075901 UP000075902 UP000076408 UP000007062 UP000075885 UP000000673 UP000069272 UP000053825 UP000092462 UP000192223 UP000075900 UP000078540 UP000078541 UP000005205 UP000075809 UP000036403 UP000075886 UP000053097 UP000007755 UP000069940 UP000249989 UP000008820 UP000078492 UP000002320 UP000075884 UP000279307 UP000079169 UP000242457 UP000005203 UP000092461 UP000075880 UP000030765 UP000015102 UP000002282 UP000015103 UP000008711 UP000037069 UP000007798 UP000268350 UP000001819 UP000008744 UP000001070 UP000078200 UP000007801 UP000009192
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JQH3
A0A2A4JBT4
A0A2H1VXB2
S4PMN4
A0A3S2NW85
A0A212ELI2
+ More
A0A194QQC4 I4DLD8 A0A0L7L117 D6X221 A0A2J7Q722 V5I7Y6 U4UKA1 A0A067R1Q5 N6SV95 J3JXB8 E2BHI5 A0A195CTQ6 R4UKZ0 A0A1B6MDB0 A0A232EYK4 K7JAQ8 A0A2S2NGK1 C4WSI6 A0A0V0G5R6 A0A224XZW6 A0A0T6B6I4 A0A1L8DU37 A0A0P4VK23 U5EZA6 T1DHD4 A0A2M3Z6Z5 A0A1B6HCG7 A0A182WWI6 A0A182UPF3 A0A182JRP3 A0A182I6R7 A0A0N0U709 A0A182MCN7 A0A182WBI9 A0A182SL99 A0A182UDD8 A0A1Q3FBZ0 A0A182Y2C2 Q7QIM0 A0A182PII6 A0A1B6G7J4 R4WT38 W5JV23 A0A182FEL8 A0A2M4C2Z2 A0A0L7QQ59 A0A1B0D5V5 A0A1W4WHZ9 A0A2M4AGV2 A0A182RJL6 A0A195BP16 A0A195FR15 A0A158NTS0 A0A151WX69 A0A0J7NNA4 A0A182QFC1 A0A026W939 F4WJ11 A0A182HA99 Q17IJ1 A0A151J594 B0W5L5 A0A151JNR5 A0A0K8TP25 A0A182N9A1 A0A1Y1KJW0 A0A3L8DA49 A0A1S3DCZ7 A0A2A3EG24 V9ILX5 A0A088ATX6 A0A1B0GII4 A0A182JEN9 A0A336N5J2 A0A1B6DQ76 A0A084W148 T1GKH0 T1J251 B4PIT1 A0A336L8G3 T1HUH4 B3NGB1 A0A0L0CCD3 W8B6K5 B4MM11 A0A3B0JW25 Q2LZL9 B4H3B9 A0A0K8UTS8 A0A034VEH7 B4IX17 A0A1A9UP26 B3M3Q7 B4KXE6
A0A194QQC4 I4DLD8 A0A0L7L117 D6X221 A0A2J7Q722 V5I7Y6 U4UKA1 A0A067R1Q5 N6SV95 J3JXB8 E2BHI5 A0A195CTQ6 R4UKZ0 A0A1B6MDB0 A0A232EYK4 K7JAQ8 A0A2S2NGK1 C4WSI6 A0A0V0G5R6 A0A224XZW6 A0A0T6B6I4 A0A1L8DU37 A0A0P4VK23 U5EZA6 T1DHD4 A0A2M3Z6Z5 A0A1B6HCG7 A0A182WWI6 A0A182UPF3 A0A182JRP3 A0A182I6R7 A0A0N0U709 A0A182MCN7 A0A182WBI9 A0A182SL99 A0A182UDD8 A0A1Q3FBZ0 A0A182Y2C2 Q7QIM0 A0A182PII6 A0A1B6G7J4 R4WT38 W5JV23 A0A182FEL8 A0A2M4C2Z2 A0A0L7QQ59 A0A1B0D5V5 A0A1W4WHZ9 A0A2M4AGV2 A0A182RJL6 A0A195BP16 A0A195FR15 A0A158NTS0 A0A151WX69 A0A0J7NNA4 A0A182QFC1 A0A026W939 F4WJ11 A0A182HA99 Q17IJ1 A0A151J594 B0W5L5 A0A151JNR5 A0A0K8TP25 A0A182N9A1 A0A1Y1KJW0 A0A3L8DA49 A0A1S3DCZ7 A0A2A3EG24 V9ILX5 A0A088ATX6 A0A1B0GII4 A0A182JEN9 A0A336N5J2 A0A1B6DQ76 A0A084W148 T1GKH0 T1J251 B4PIT1 A0A336L8G3 T1HUH4 B3NGB1 A0A0L0CCD3 W8B6K5 B4MM11 A0A3B0JW25 Q2LZL9 B4H3B9 A0A0K8UTS8 A0A034VEH7 B4IX17 A0A1A9UP26 B3M3Q7 B4KXE6
Ontologies
GO
PANTHER
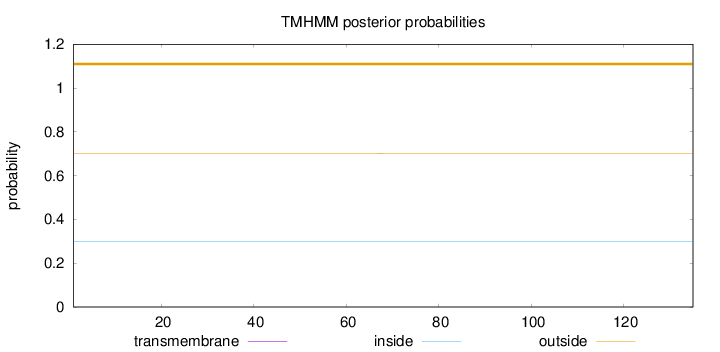
Topology
Length:
135
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00127
Exp number, first 60 AAs:
0
Total prob of N-in:
0.29926
outside
1 - 135
Population Genetic Test Statistics
Pi
147.509206
Theta
147.374144
Tajima's D
-0.798856
CLR
468.522874
CSRT
0.175791210439478
Interpretation
Uncertain
