Gene
KWMTBOMO06642
Pre Gene Modal
BGIBMGA011700
Annotation
PREDICTED:_sodium/hydrogen_exchanger_8_[Papilio_xuthus]
Full name
Sodium/hydrogen exchanger
Location in the cell
PlasmaMembrane Reliability : 4.968
Sequence
CDS
ATGAATGTTAATCAAACAGTCGATTCAACAACTCCAAGTTCAACCGAAAAATCGAATATAGTTTACGAAATAGGACCATCAAATTCTTCAATACTGGCAGTAGTTCAAGTTAATGCTTCGGCTGCGACACCCAATGCCTCTGTTAATCCATCAGACGCTGCTCCTCTTCCAGAAAAAAGCTCAGCTGAAGATGAACATAATAGCTCAATGGCAATATTTTTCGTACTCTGTATTTTGGCCTTAGGTATACTGTTAATACATTTAATGTTACAAACAGGATTTTCTTATCTCCCTGAAAGTGTGGTTATAGTATTTCTTGGTGCTCTCATTGGAATGATAATAAATCTTATGTCGATAAAAAATATTGCCAATTGGAGAAAGGAAGAAGCATTTTCACCAACAGCTTTCTTTTTAGTGTTGCTACCACCCATCATATTTGAATCGGGTTATAATCTCCACAAAGGCAACTTTTTTCAAAATATTGGTTCAATATTGTTATTTGCTATCGTAGGAACTGCAATATCAGCATTTGTAATTGGAGCTGGTATCTACTTATTGGGTTTGGCTCAGGTTGTATATAAACTTAGCTTTGTTGAGTCATTTGCTTTTGGATCCCTGATATCTGCTGTGGATCCAGTCGCCACTGTGGCTATATTTCACGCTCTAGATGTTGACCCAGTTCTCAACATGTTGGTATTTGGGGAGAGTATATTAAATGATGCTGTGGCTATTGTGTTGACAACAGCCGTTCTGGATTCCAATGACCCTTTCATGTCTACAGGAGAGGCAATTCTCTCTGCAATTAATCGATTTTGGTTAATGTTCTTTGCATCAGCTGGCATTGGTGTACTTTTTGCACTTGTGAGTGCCTTACTCTTGAAACATGTTGATCTCAGAAAGAATCCATCTCTTGAGTTTGGAATGATGTTGGTATTCACATATGCTCCTTATGTGTTAGCTGAAGGCATTCACTTGTCAGGGATTATGGCTATACTGTTTTGTGGTATTGTGATGTCTCATTACACTCATTTTAATTTATCCACAATTACTCAAGTTACAATGCAACAAACAATGAGAACTCTTGCATTCATCGCGGAAACATGTGTTTTTGCATATTTAGGACTTGCTCTATTCAGTTTCAGACATCGCGTCGAACCTGCACTGGTAGTCTGGAGTATAGTAATGTGTCTCCTTGGCAGAGCAGCTAATATATTCCCTTTGGCACTGCTTTGCAACAAATTTCGAGAACACAAGATTACCAAGAAAATGATGTTTATTATGTGGTTCAGCGGTCTAAGGGGTGCGATATCTTACGCTTTATCTCTGCATCTTGGATTTTCGGATGAAACCAGACATGTGATTATAACAACGACACTCATAATCGTGTTGTTTACTACTTTGTTCTTCGGAGGGTCCACTATGCCACTATTGAAATTTTTACGGGCAAACAAGAAAACAAAACGTGCACGACCTTTGCGCAAACAAAAGGAAGTAAGTTTGTCTAAAACAAAAGAGTGGGGGCAAGCAATCGATTCTGAACATTTATCTGAAATCACTGAAGAGGAACTTGAGGTGAATTATTCACACGCAACTCGTTTACGGGGCTTCGTACGATTTGATATGAAATATCTGACCCCGTTTTTTACGCGACGGTTTACTCATCAGGAGCTAAAAGATTGCAAAAGCCAAATGACAGACCTGACAAATCAATGGTATCAAGCGATTAGGATATCCAATGATAGAGAAACGGATGAAGAAGAAATTTTACAGCAGAATTCCCTTCCGAGCTCACGCAATAGTCTGCAAAGAGGTGTTTAG
Protein
MNVNQTVDSTTPSSTEKSNIVYEIGPSNSSILAVVQVNASAATPNASVNPSDAAPLPEKSSAEDEHNSSMAIFFVLCILALGILLIHLMLQTGFSYLPESVVIVFLGALIGMIINLMSIKNIANWRKEEAFSPTAFFLVLLPPIIFESGYNLHKGNFFQNIGSILLFAIVGTAISAFVIGAGIYLLGLAQVVYKLSFVESFAFGSLISAVDPVATVAIFHALDVDPVLNMLVFGESILNDAVAIVLTTAVLDSNDPFMSTGEAILSAINRFWLMFFASAGIGVLFALVSALLLKHVDLRKNPSLEFGMMLVFTYAPYVLAEGIHLSGIMAILFCGIVMSHYTHFNLSTITQVTMQQTMRTLAFIAETCVFAYLGLALFSFRHRVEPALVVWSIVMCLLGRAANIFPLALLCNKFREHKITKKMMFIMWFSGLRGAISYALSLHLGFSDETRHVIITTTLIIVLFTTLFFGGSTMPLLKFLRANKKTKRARPLRKQKEVSLSKTKEWGQAIDSEHLSEITEEELEVNYSHATRLRGFVRFDMKYLTPFFTRRFTHQELKDCKSQMTDLTNQWYQAIRISNDRETDEEEILQQNSLPSSRNSLQRGV
Summary
Similarity
Belongs to the monovalent cation:proton antiporter 1 (CPA1) transporter (TC 2.A.36) family.
Belongs to the DPH1/DPH2 family.
Belongs to the DPH1/DPH2 family.
Feature
chain Sodium/hydrogen exchanger
Uniprot
A0A194PX96
A0A2Z4GWW3
A0A2A4JBD8
A9QVL3
A0A2H1VX86
A0A3S2NFH9
+ More
A0A194QUD1 A0A212FDD6 A0A151WNV1 A0A2J7QB86 A0A2J7QB85 A0A0T6B3G6 E2B5T7 A0A0C9QG15 E2A1X6 A0A0C9RJM7 A0A151IIS8 E9IGJ8 A0A0J7KY84 A0A026W8M9 A0A3L8DRK0 A0A224XCE4 A0A2A3EIR0 A0A195BDL0 A0A151JYA3 A0A023F4U9 A0A195DPC5 A0A158NFI9 A0A2P8XM38 A0A146KTU6 A0A232FLC6 K7IQS2 A0A1B6L8B6 A0A1B6GTN8 A0A1W4WR07 A0A1B6CIL1 A0A2S2NUE7 A0A2H8TYV5 J9JJV7 A0A2S2R391 A0A1Y1LJ00 A0A310SEU4 A0A087ZZD1 A0A0L7L120 A0A0L7R952 F4X190 A0A0P4VFZ1 E0VLV5 U4UA95 N6TPG0 A0A067R5K9 A0A1S4F7V9 Q4L225 A0A182GWA7 A0A1L8DUX0 A0A1L8DUW5 A0A023EUT3 B0W243 A0A1L8DUW2 B0X6D4 A0A1Q3FER8 A0A1Q3FBV6 A0A1Q3FEV4 A0A1Q3FBQ6 V9IC16 T1PKQ7 A0A1I8PAH8 Q7QKG3 A0A0L0BLR8 V5GSC0 A0A182PJ05 A0A182HG44 A0A182V7N2 T1E186 A0A182XIP3 A0A1S4H161 A0A2M4BFS5 W5J8T6 A0A182TDT8 A0A2M3Z2U6 A0A2M3ZE89 A0A2M4A788 A0A182F5A4 A0A182K1L7 A0A182M4P3 A0A182JBD2 A0A182VUT8 A0A1A9W0H6 A0A182RGA3 A0A1A9V8X5 W8BLB4 A0A0K8TYZ1 A0A182QT37 A0A084VPS0 A0A1B0G0I4 A0A182YFZ2 A0A1A9ZJ05 A0A182NEK6 A0A034VCN8 A0A1B6ICY6
A0A194QUD1 A0A212FDD6 A0A151WNV1 A0A2J7QB86 A0A2J7QB85 A0A0T6B3G6 E2B5T7 A0A0C9QG15 E2A1X6 A0A0C9RJM7 A0A151IIS8 E9IGJ8 A0A0J7KY84 A0A026W8M9 A0A3L8DRK0 A0A224XCE4 A0A2A3EIR0 A0A195BDL0 A0A151JYA3 A0A023F4U9 A0A195DPC5 A0A158NFI9 A0A2P8XM38 A0A146KTU6 A0A232FLC6 K7IQS2 A0A1B6L8B6 A0A1B6GTN8 A0A1W4WR07 A0A1B6CIL1 A0A2S2NUE7 A0A2H8TYV5 J9JJV7 A0A2S2R391 A0A1Y1LJ00 A0A310SEU4 A0A087ZZD1 A0A0L7L120 A0A0L7R952 F4X190 A0A0P4VFZ1 E0VLV5 U4UA95 N6TPG0 A0A067R5K9 A0A1S4F7V9 Q4L225 A0A182GWA7 A0A1L8DUX0 A0A1L8DUW5 A0A023EUT3 B0W243 A0A1L8DUW2 B0X6D4 A0A1Q3FER8 A0A1Q3FBV6 A0A1Q3FEV4 A0A1Q3FBQ6 V9IC16 T1PKQ7 A0A1I8PAH8 Q7QKG3 A0A0L0BLR8 V5GSC0 A0A182PJ05 A0A182HG44 A0A182V7N2 T1E186 A0A182XIP3 A0A1S4H161 A0A2M4BFS5 W5J8T6 A0A182TDT8 A0A2M3Z2U6 A0A2M3ZE89 A0A2M4A788 A0A182F5A4 A0A182K1L7 A0A182M4P3 A0A182JBD2 A0A182VUT8 A0A1A9W0H6 A0A182RGA3 A0A1A9V8X5 W8BLB4 A0A0K8TYZ1 A0A182QT37 A0A084VPS0 A0A1B0G0I4 A0A182YFZ2 A0A1A9ZJ05 A0A182NEK6 A0A034VCN8 A0A1B6ICY6
Pubmed
26354079
29908900
22118469
20798317
21282665
24508170
+ More
30249741 25474469 21347285 29403074 26823975 28648823 20075255 28004739 26227816 21719571 27129103 20566863 23537049 24845553 17510324 19193723 26483478 24945155 25315136 12364791 26108605 20920257 23761445 24495485 24438588 25244985 25348373
30249741 25474469 21347285 29403074 26823975 28648823 20075255 28004739 26227816 21719571 27129103 20566863 23537049 24845553 17510324 19193723 26483478 24945155 25315136 12364791 26108605 20920257 23761445 24495485 24438588 25244985 25348373
EMBL
KQ459590
KPI97384.1
MH048893
AWW06965.1
NWSH01002234
PCG68864.1
+ More
EU272023 ABX71221.2 ODYU01005003 SOQ45449.1 RSAL01000137 RVE46194.1 KQ461190 KPJ07161.1 AGBW02009074 OWR51751.1 KQ982893 KYQ49582.1 NEVH01016301 PNF25845.1 PNF25844.1 LJIG01016014 KRT81874.1 GL445887 EFN88906.1 GBYB01013468 JAG83235.1 GL435845 EFN72570.1 GBYB01013467 JAG83234.1 KQ977464 KYN02550.1 GL763048 EFZ20298.1 LBMM01001942 KMQ95502.1 KK107361 EZA51996.1 QOIP01000005 RLU23064.1 GFTR01007782 JAW08644.1 KZ288230 PBC31578.1 KQ976514 KYM82292.1 KQ981494 KYN40946.1 GBBI01002216 JAC16496.1 KQ980662 KYN14696.1 ADTU01014526 PYGN01001739 PSN33069.1 GDHC01018718 JAP99910.1 NNAY01000082 OXU31147.1 GEBQ01020028 JAT19949.1 GECZ01003992 JAS65777.1 GEDC01024010 JAS13288.1 GGMR01008043 MBY20662.1 GFXV01006533 MBW18338.1 ABLF02039927 GGMS01015191 MBY84394.1 GEZM01054490 JAV73613.1 KQ767065 OAD53516.1 JTDY01003664 KOB69193.1 KQ414627 KOC67400.1 GL888528 EGI59762.1 GDKW01003583 JAI53012.1 DS235281 EEB14361.1 KB632195 ERL89967.1 APGK01026647 KB740635 ENN79908.1 KK852729 KDR17572.1 AY326255 EU760347 CH477306 AAQ91612.1 ACJ02512.1 EAT44116.1 JXUM01092896 JXUM01092897 JXUM01092898 JXUM01092899 KQ564018 KXJ72926.1 GFDF01003861 JAV10223.1 GFDF01003862 JAV10222.1 GAPW01000403 JAC13195.1 DS231825 EDS28171.1 GFDF01003863 JAV10221.1 DS232411 EDS41284.1 GFDL01009001 JAV26044.1 GFDL01010008 JAV25037.1 GFDL01008993 JAV26052.1 GFDL01010034 JAV25011.1 JR039365 AEY58658.1 KA648523 AFP63152.1 AAAB01008797 EAA03626.4 JRES01001684 KNC20972.1 GALX01003974 JAB64492.1 APCN01004115 GAKT01000109 JAA92953.1 GGFJ01002765 MBW51906.1 ADMH02002093 ETN59290.1 GGFM01002093 MBW22844.1 GGFM01006072 MBW26823.1 GGFK01003290 MBW36611.1 AXCM01000484 AXCP01008030 GAMC01006988 JAB99567.1 GDHF01032813 JAI19501.1 AXCN02000851 ATLV01015034 KE524999 KFB39964.1 CCAG010017281 GAKP01019070 GAKP01019068 GAKP01019066 JAC39882.1 GECU01022921 JAS84785.1
EU272023 ABX71221.2 ODYU01005003 SOQ45449.1 RSAL01000137 RVE46194.1 KQ461190 KPJ07161.1 AGBW02009074 OWR51751.1 KQ982893 KYQ49582.1 NEVH01016301 PNF25845.1 PNF25844.1 LJIG01016014 KRT81874.1 GL445887 EFN88906.1 GBYB01013468 JAG83235.1 GL435845 EFN72570.1 GBYB01013467 JAG83234.1 KQ977464 KYN02550.1 GL763048 EFZ20298.1 LBMM01001942 KMQ95502.1 KK107361 EZA51996.1 QOIP01000005 RLU23064.1 GFTR01007782 JAW08644.1 KZ288230 PBC31578.1 KQ976514 KYM82292.1 KQ981494 KYN40946.1 GBBI01002216 JAC16496.1 KQ980662 KYN14696.1 ADTU01014526 PYGN01001739 PSN33069.1 GDHC01018718 JAP99910.1 NNAY01000082 OXU31147.1 GEBQ01020028 JAT19949.1 GECZ01003992 JAS65777.1 GEDC01024010 JAS13288.1 GGMR01008043 MBY20662.1 GFXV01006533 MBW18338.1 ABLF02039927 GGMS01015191 MBY84394.1 GEZM01054490 JAV73613.1 KQ767065 OAD53516.1 JTDY01003664 KOB69193.1 KQ414627 KOC67400.1 GL888528 EGI59762.1 GDKW01003583 JAI53012.1 DS235281 EEB14361.1 KB632195 ERL89967.1 APGK01026647 KB740635 ENN79908.1 KK852729 KDR17572.1 AY326255 EU760347 CH477306 AAQ91612.1 ACJ02512.1 EAT44116.1 JXUM01092896 JXUM01092897 JXUM01092898 JXUM01092899 KQ564018 KXJ72926.1 GFDF01003861 JAV10223.1 GFDF01003862 JAV10222.1 GAPW01000403 JAC13195.1 DS231825 EDS28171.1 GFDF01003863 JAV10221.1 DS232411 EDS41284.1 GFDL01009001 JAV26044.1 GFDL01010008 JAV25037.1 GFDL01008993 JAV26052.1 GFDL01010034 JAV25011.1 JR039365 AEY58658.1 KA648523 AFP63152.1 AAAB01008797 EAA03626.4 JRES01001684 KNC20972.1 GALX01003974 JAB64492.1 APCN01004115 GAKT01000109 JAA92953.1 GGFJ01002765 MBW51906.1 ADMH02002093 ETN59290.1 GGFM01002093 MBW22844.1 GGFM01006072 MBW26823.1 GGFK01003290 MBW36611.1 AXCM01000484 AXCP01008030 GAMC01006988 JAB99567.1 GDHF01032813 JAI19501.1 AXCN02000851 ATLV01015034 KE524999 KFB39964.1 CCAG010017281 GAKP01019070 GAKP01019068 GAKP01019066 JAC39882.1 GECU01022921 JAS84785.1
Proteomes
UP000053268
UP000218220
UP000283053
UP000053240
UP000007151
UP000075809
+ More
UP000235965 UP000008237 UP000000311 UP000078542 UP000036403 UP000053097 UP000279307 UP000242457 UP000078540 UP000078541 UP000078492 UP000005205 UP000245037 UP000215335 UP000002358 UP000192223 UP000007819 UP000005203 UP000037510 UP000053825 UP000007755 UP000009046 UP000030742 UP000019118 UP000027135 UP000008820 UP000069940 UP000249989 UP000002320 UP000095301 UP000095300 UP000007062 UP000037069 UP000075885 UP000075840 UP000075903 UP000076407 UP000000673 UP000075902 UP000069272 UP000075881 UP000075883 UP000075880 UP000075920 UP000091820 UP000075900 UP000078200 UP000075886 UP000030765 UP000092444 UP000076408 UP000092445 UP000075884
UP000235965 UP000008237 UP000000311 UP000078542 UP000036403 UP000053097 UP000279307 UP000242457 UP000078540 UP000078541 UP000078492 UP000005205 UP000245037 UP000215335 UP000002358 UP000192223 UP000007819 UP000005203 UP000037510 UP000053825 UP000007755 UP000009046 UP000030742 UP000019118 UP000027135 UP000008820 UP000069940 UP000249989 UP000002320 UP000095301 UP000095300 UP000007062 UP000037069 UP000075885 UP000075840 UP000075903 UP000076407 UP000000673 UP000075902 UP000069272 UP000075881 UP000075883 UP000075880 UP000075920 UP000091820 UP000075900 UP000078200 UP000075886 UP000030765 UP000092444 UP000076408 UP000092445 UP000075884
Interpro
Gene 3D
ProteinModelPortal
A0A194PX96
A0A2Z4GWW3
A0A2A4JBD8
A9QVL3
A0A2H1VX86
A0A3S2NFH9
+ More
A0A194QUD1 A0A212FDD6 A0A151WNV1 A0A2J7QB86 A0A2J7QB85 A0A0T6B3G6 E2B5T7 A0A0C9QG15 E2A1X6 A0A0C9RJM7 A0A151IIS8 E9IGJ8 A0A0J7KY84 A0A026W8M9 A0A3L8DRK0 A0A224XCE4 A0A2A3EIR0 A0A195BDL0 A0A151JYA3 A0A023F4U9 A0A195DPC5 A0A158NFI9 A0A2P8XM38 A0A146KTU6 A0A232FLC6 K7IQS2 A0A1B6L8B6 A0A1B6GTN8 A0A1W4WR07 A0A1B6CIL1 A0A2S2NUE7 A0A2H8TYV5 J9JJV7 A0A2S2R391 A0A1Y1LJ00 A0A310SEU4 A0A087ZZD1 A0A0L7L120 A0A0L7R952 F4X190 A0A0P4VFZ1 E0VLV5 U4UA95 N6TPG0 A0A067R5K9 A0A1S4F7V9 Q4L225 A0A182GWA7 A0A1L8DUX0 A0A1L8DUW5 A0A023EUT3 B0W243 A0A1L8DUW2 B0X6D4 A0A1Q3FER8 A0A1Q3FBV6 A0A1Q3FEV4 A0A1Q3FBQ6 V9IC16 T1PKQ7 A0A1I8PAH8 Q7QKG3 A0A0L0BLR8 V5GSC0 A0A182PJ05 A0A182HG44 A0A182V7N2 T1E186 A0A182XIP3 A0A1S4H161 A0A2M4BFS5 W5J8T6 A0A182TDT8 A0A2M3Z2U6 A0A2M3ZE89 A0A2M4A788 A0A182F5A4 A0A182K1L7 A0A182M4P3 A0A182JBD2 A0A182VUT8 A0A1A9W0H6 A0A182RGA3 A0A1A9V8X5 W8BLB4 A0A0K8TYZ1 A0A182QT37 A0A084VPS0 A0A1B0G0I4 A0A182YFZ2 A0A1A9ZJ05 A0A182NEK6 A0A034VCN8 A0A1B6ICY6
A0A194QUD1 A0A212FDD6 A0A151WNV1 A0A2J7QB86 A0A2J7QB85 A0A0T6B3G6 E2B5T7 A0A0C9QG15 E2A1X6 A0A0C9RJM7 A0A151IIS8 E9IGJ8 A0A0J7KY84 A0A026W8M9 A0A3L8DRK0 A0A224XCE4 A0A2A3EIR0 A0A195BDL0 A0A151JYA3 A0A023F4U9 A0A195DPC5 A0A158NFI9 A0A2P8XM38 A0A146KTU6 A0A232FLC6 K7IQS2 A0A1B6L8B6 A0A1B6GTN8 A0A1W4WR07 A0A1B6CIL1 A0A2S2NUE7 A0A2H8TYV5 J9JJV7 A0A2S2R391 A0A1Y1LJ00 A0A310SEU4 A0A087ZZD1 A0A0L7L120 A0A0L7R952 F4X190 A0A0P4VFZ1 E0VLV5 U4UA95 N6TPG0 A0A067R5K9 A0A1S4F7V9 Q4L225 A0A182GWA7 A0A1L8DUX0 A0A1L8DUW5 A0A023EUT3 B0W243 A0A1L8DUW2 B0X6D4 A0A1Q3FER8 A0A1Q3FBV6 A0A1Q3FEV4 A0A1Q3FBQ6 V9IC16 T1PKQ7 A0A1I8PAH8 Q7QKG3 A0A0L0BLR8 V5GSC0 A0A182PJ05 A0A182HG44 A0A182V7N2 T1E186 A0A182XIP3 A0A1S4H161 A0A2M4BFS5 W5J8T6 A0A182TDT8 A0A2M3Z2U6 A0A2M3ZE89 A0A2M4A788 A0A182F5A4 A0A182K1L7 A0A182M4P3 A0A182JBD2 A0A182VUT8 A0A1A9W0H6 A0A182RGA3 A0A1A9V8X5 W8BLB4 A0A0K8TYZ1 A0A182QT37 A0A084VPS0 A0A1B0G0I4 A0A182YFZ2 A0A1A9ZJ05 A0A182NEK6 A0A034VCN8 A0A1B6ICY6
PDB
4CZ9
E-value=2.51596e-11,
Score=168
Ontologies
GO
PANTHER
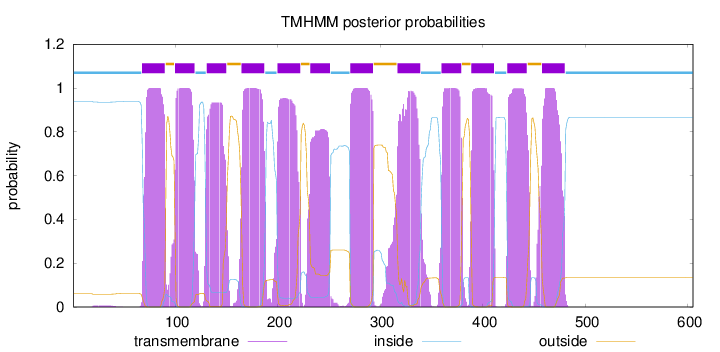
Topology
Length:
605
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
261.11126
Exp number, first 60 AAs:
0.1382
Total prob of N-in:
0.93958
inside
1 - 67
TMhelix
68 - 90
outside
91 - 99
TMhelix
100 - 119
inside
120 - 130
TMhelix
131 - 150
outside
151 - 164
TMhelix
165 - 187
inside
188 - 199
TMhelix
200 - 222
outside
223 - 231
TMhelix
232 - 251
inside
252 - 270
TMhelix
271 - 293
outside
294 - 316
TMhelix
317 - 339
inside
340 - 359
TMhelix
360 - 379
outside
380 - 388
TMhelix
389 - 411
inside
412 - 423
TMhelix
424 - 443
outside
444 - 457
TMhelix
458 - 480
inside
481 - 605
Population Genetic Test Statistics
Pi
208.043546
Theta
20.551157
Tajima's D
-1.412659
CLR
201.284344
CSRT
0.0689465526723664
Interpretation
Uncertain
