Gene
KWMTBOMO06636 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011698
Annotation
PREDICTED:_ribonuclease_UK114-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 0.718
Sequence
CDS
ATGAGCTCAGAAGATTCTGCACTGGCGTCCGAAGACAAAGTTTCGCCCAAAATGAGTTCAGCAAAAAATTCGAATAACAATAAAGTAACAAAAACAATAATAACATCTCCGGAAATATATCAACCAGTAGGTCCGTACAGTCAAGCAATTTTAGCGGACAAGACCTTATACATTTCTGGAATTCTCGGATTGGATCGAGATGCACAGATGGTCTGCGGTGGTGCTGAAGCGCAGACCCGTCAGGCTCTGGACAATCTGCGACATGTACTTGAAGCTGGTGGCGCTTCGTTGGAGTCGGTCGTGAAAACTACTGTTTTGTTGGCTAGCATGGACGACTTCCAAACTTTCAACAAAGTCTATGCAGAATATTTTCCTAAAGCTTGCCCTGCTCGAATGACATACGAAGTCAGTCGACTACCGTTGGGAGCAGCTGTGGAGATCGAGGCCATCGCTCTTTGCGGAGACTTGGTCATAGCAGAGGCCGGCCCATGCCCTTGTGCACGCTAA
Protein
MSSEDSALASEDKVSPKMSSAKNSNNNKVTKTIITSPEIYQPVGPYSQAILADKTLYISGILGLDRDAQMVCGGAEAQTRQALDNLRHVLEAGGASLESVVKTTVLLASMDDFQTFNKVYAEYFPKACPARMTYEVSRLPLGAAVEIEAIALCGDLVIAEAGPCPCAR
Summary
Uniprot
H9JQ92
A0A437AV15
A0A0L7LL19
A0A212FDB5
A0A2A4JF56
S4PA09
+ More
A0A212FFK2 A0A2W1B4V0 A0A2A4JYS6 A0A2H1X0W4 A0A0L7KL57 A0A194PVM6 A0A194QNS5 D6WVK4 T1II18 A0A1Y4AX62 A0A088A6M8 A0A034WU14 A0A0C9QHR2 A0A437CHR9 A0A336MQ35 A0A0K8U816 A0A0T6B8H6 A0A1Y1JU25 A0A077Z8Z3 A0A1Y1JZI9 A0A2C9KLV8 A0A2A3EEU2 A0A3M1M8S6 A0A3P9JBI1 A0A224XWA0 C1BRB9 A0A3P9HVP1 A0A1X7UCT0 A0A069DVY8 A0A146MVE7 A0A3Q3EIB4 A0A146W0H0 A0A0V0GBM8 A0A3B3TSC8 A0A3M1MHJ1 A0A3B3IFE4 C1BQE7 A0A0P4VJC4 R4G3R0 A0A2K6F0A1 A0A336M697 A0A2I4CG74 C3KJM3 H0WIN4 A0A085MAB3 A0A1W5AY64 A0A146ZXA2 A0A0S7HFE7 A0A3B3XP86 A0A3B5MTJ1 A0A3B4BGB0 E0VAS8 A0A226E348 A0A131Z494 G3MSD3 A0A067QY57 A0A3B5AHI7 C3KH08 A0A3B4TWR5 A0A3B3D813 A0A131XL15 A0A3Q2DIC4 A0A3B1KHR4 A0A1A7XWU2 A0A3B4F597 A0A3P9DIR8 A0A3P8SEV9 G5B8Y4 A0A3Q4GIG3 I3MHC8 I3KHW6 A0A1H4EH84 A0A3B5QXT5 G7PHN9 A0A2K6NJW1 A0A0M4EAK3 H2TRV4 A0A2K6JSX5 A0A3Q1JRQ1 A0A0B6YBB3 A0A3Q1B131 T1F395 A0A3S1B6V3 A0A2Y9JA97
A0A212FFK2 A0A2W1B4V0 A0A2A4JYS6 A0A2H1X0W4 A0A0L7KL57 A0A194PVM6 A0A194QNS5 D6WVK4 T1II18 A0A1Y4AX62 A0A088A6M8 A0A034WU14 A0A0C9QHR2 A0A437CHR9 A0A336MQ35 A0A0K8U816 A0A0T6B8H6 A0A1Y1JU25 A0A077Z8Z3 A0A1Y1JZI9 A0A2C9KLV8 A0A2A3EEU2 A0A3M1M8S6 A0A3P9JBI1 A0A224XWA0 C1BRB9 A0A3P9HVP1 A0A1X7UCT0 A0A069DVY8 A0A146MVE7 A0A3Q3EIB4 A0A146W0H0 A0A0V0GBM8 A0A3B3TSC8 A0A3M1MHJ1 A0A3B3IFE4 C1BQE7 A0A0P4VJC4 R4G3R0 A0A2K6F0A1 A0A336M697 A0A2I4CG74 C3KJM3 H0WIN4 A0A085MAB3 A0A1W5AY64 A0A146ZXA2 A0A0S7HFE7 A0A3B3XP86 A0A3B5MTJ1 A0A3B4BGB0 E0VAS8 A0A226E348 A0A131Z494 G3MSD3 A0A067QY57 A0A3B5AHI7 C3KH08 A0A3B4TWR5 A0A3B3D813 A0A131XL15 A0A3Q2DIC4 A0A3B1KHR4 A0A1A7XWU2 A0A3B4F597 A0A3P9DIR8 A0A3P8SEV9 G5B8Y4 A0A3Q4GIG3 I3MHC8 I3KHW6 A0A1H4EH84 A0A3B5QXT5 G7PHN9 A0A2K6NJW1 A0A0M4EAK3 H2TRV4 A0A2K6JSX5 A0A3Q1JRQ1 A0A0B6YBB3 A0A3Q1B131 T1F395 A0A3S1B6V3 A0A2Y9JA97
Pubmed
EMBL
BABH01011999
RSAL01000395
RVE41971.1
JTDY01000710
KOB76132.1
AGBW02009074
+ More
OWR51745.1 NWSH01001612 PCG70687.1 GAIX01006617 JAA85943.1 AGBW02008810 OWR52514.1 KZ150337 PZC71319.1 NWSH01000384 PCG76794.1 ODYU01012587 SOQ59000.1 JTDY01009347 KOB63624.1 KQ459590 KPI97377.1 KQ461190 KPJ07167.1 KQ971357 EFA08585.1 JH430074 NFIO01000001 OUO23421.1 GAKP01000823 JAC58129.1 GBYB01000107 JAG69874.1 CM012452 RVE62195.1 UFQT01001742 SSX31621.1 GDHF01029520 JAI22794.1 LJIG01009150 KRT83656.1 GEZM01100711 JAV52814.1 HG806020 CDW56234.1 GEZM01100710 JAV52815.1 KZ288263 PBC30303.1 RFIW01000015 RMF38973.1 GFTR01002278 JAW14148.1 BT077148 ACO11572.1 GBGD01003420 JAC85469.1 GCES01162823 JAQ23499.1 GCES01063238 JAR23085.1 GECL01000709 JAP05415.1 RFIS01000730 RMF42998.1 BT076826 ACO11250.1 GDKW01001517 JAI55078.1 ACPB03013485 GAHY01001446 JAA76064.1 UFQT01000446 SSX24409.1 BT083138 ACQ58845.1 AAQR03010895 AAQR03010896 AAQR03010897 AAQR03010898 KL363210 KFD54159.1 GCES01015369 JAR70954.1 GBYX01441561 JAO39815.1 DS235015 EEB10484.1 LNIX01000007 OXA51397.1 GEDV01003291 JAP85266.1 JO844784 AEO36401.1 KK853119 KDR11138.1 BT082223 ACQ57930.1 GEFH01000738 JAP67843.1 HADW01010387 HADX01000008 SBP22240.1 JH169014 GEBF01005362 EHB05745.1 JAN98270.1 AGTP01002012 AERX01058199 FNRI01000007 SEA83950.1 CM001286 EHH66215.1 CP012523 ALC38260.1 HACG01006533 CEK53398.1 AMQM01003615 KB096222 ESO07303.1 RQTK01000665 RUS76402.1
OWR51745.1 NWSH01001612 PCG70687.1 GAIX01006617 JAA85943.1 AGBW02008810 OWR52514.1 KZ150337 PZC71319.1 NWSH01000384 PCG76794.1 ODYU01012587 SOQ59000.1 JTDY01009347 KOB63624.1 KQ459590 KPI97377.1 KQ461190 KPJ07167.1 KQ971357 EFA08585.1 JH430074 NFIO01000001 OUO23421.1 GAKP01000823 JAC58129.1 GBYB01000107 JAG69874.1 CM012452 RVE62195.1 UFQT01001742 SSX31621.1 GDHF01029520 JAI22794.1 LJIG01009150 KRT83656.1 GEZM01100711 JAV52814.1 HG806020 CDW56234.1 GEZM01100710 JAV52815.1 KZ288263 PBC30303.1 RFIW01000015 RMF38973.1 GFTR01002278 JAW14148.1 BT077148 ACO11572.1 GBGD01003420 JAC85469.1 GCES01162823 JAQ23499.1 GCES01063238 JAR23085.1 GECL01000709 JAP05415.1 RFIS01000730 RMF42998.1 BT076826 ACO11250.1 GDKW01001517 JAI55078.1 ACPB03013485 GAHY01001446 JAA76064.1 UFQT01000446 SSX24409.1 BT083138 ACQ58845.1 AAQR03010895 AAQR03010896 AAQR03010897 AAQR03010898 KL363210 KFD54159.1 GCES01015369 JAR70954.1 GBYX01441561 JAO39815.1 DS235015 EEB10484.1 LNIX01000007 OXA51397.1 GEDV01003291 JAP85266.1 JO844784 AEO36401.1 KK853119 KDR11138.1 BT082223 ACQ57930.1 GEFH01000738 JAP67843.1 HADW01010387 HADX01000008 SBP22240.1 JH169014 GEBF01005362 EHB05745.1 JAN98270.1 AGTP01002012 AERX01058199 FNRI01000007 SEA83950.1 CM001286 EHH66215.1 CP012523 ALC38260.1 HACG01006533 CEK53398.1 AMQM01003615 KB096222 ESO07303.1 RQTK01000665 RUS76402.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000007151
UP000218220
UP000053268
+ More
UP000053240 UP000007266 UP000196483 UP000005203 UP000030665 UP000076420 UP000242457 UP000281227 UP000265200 UP000265180 UP000007879 UP000264800 UP000265000 UP000261500 UP000278570 UP000001038 UP000015103 UP000233160 UP000192220 UP000005225 UP000192224 UP000261480 UP000261380 UP000261520 UP000009046 UP000198287 UP000027135 UP000261400 UP000261420 UP000261560 UP000265020 UP000018467 UP000261460 UP000265160 UP000265080 UP000006813 UP000261580 UP000005215 UP000005207 UP000183253 UP000002852 UP000009130 UP000233200 UP000092553 UP000005226 UP000233180 UP000265040 UP000257160 UP000015101 UP000271974 UP000248482
UP000053240 UP000007266 UP000196483 UP000005203 UP000030665 UP000076420 UP000242457 UP000281227 UP000265200 UP000265180 UP000007879 UP000264800 UP000265000 UP000261500 UP000278570 UP000001038 UP000015103 UP000233160 UP000192220 UP000005225 UP000192224 UP000261480 UP000261380 UP000261520 UP000009046 UP000198287 UP000027135 UP000261400 UP000261420 UP000261560 UP000265020 UP000018467 UP000261460 UP000265160 UP000265080 UP000006813 UP000261580 UP000005215 UP000005207 UP000183253 UP000002852 UP000009130 UP000233200 UP000092553 UP000005226 UP000233180 UP000265040 UP000257160 UP000015101 UP000271974 UP000248482
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JQ92
A0A437AV15
A0A0L7LL19
A0A212FDB5
A0A2A4JF56
S4PA09
+ More
A0A212FFK2 A0A2W1B4V0 A0A2A4JYS6 A0A2H1X0W4 A0A0L7KL57 A0A194PVM6 A0A194QNS5 D6WVK4 T1II18 A0A1Y4AX62 A0A088A6M8 A0A034WU14 A0A0C9QHR2 A0A437CHR9 A0A336MQ35 A0A0K8U816 A0A0T6B8H6 A0A1Y1JU25 A0A077Z8Z3 A0A1Y1JZI9 A0A2C9KLV8 A0A2A3EEU2 A0A3M1M8S6 A0A3P9JBI1 A0A224XWA0 C1BRB9 A0A3P9HVP1 A0A1X7UCT0 A0A069DVY8 A0A146MVE7 A0A3Q3EIB4 A0A146W0H0 A0A0V0GBM8 A0A3B3TSC8 A0A3M1MHJ1 A0A3B3IFE4 C1BQE7 A0A0P4VJC4 R4G3R0 A0A2K6F0A1 A0A336M697 A0A2I4CG74 C3KJM3 H0WIN4 A0A085MAB3 A0A1W5AY64 A0A146ZXA2 A0A0S7HFE7 A0A3B3XP86 A0A3B5MTJ1 A0A3B4BGB0 E0VAS8 A0A226E348 A0A131Z494 G3MSD3 A0A067QY57 A0A3B5AHI7 C3KH08 A0A3B4TWR5 A0A3B3D813 A0A131XL15 A0A3Q2DIC4 A0A3B1KHR4 A0A1A7XWU2 A0A3B4F597 A0A3P9DIR8 A0A3P8SEV9 G5B8Y4 A0A3Q4GIG3 I3MHC8 I3KHW6 A0A1H4EH84 A0A3B5QXT5 G7PHN9 A0A2K6NJW1 A0A0M4EAK3 H2TRV4 A0A2K6JSX5 A0A3Q1JRQ1 A0A0B6YBB3 A0A3Q1B131 T1F395 A0A3S1B6V3 A0A2Y9JA97
A0A212FFK2 A0A2W1B4V0 A0A2A4JYS6 A0A2H1X0W4 A0A0L7KL57 A0A194PVM6 A0A194QNS5 D6WVK4 T1II18 A0A1Y4AX62 A0A088A6M8 A0A034WU14 A0A0C9QHR2 A0A437CHR9 A0A336MQ35 A0A0K8U816 A0A0T6B8H6 A0A1Y1JU25 A0A077Z8Z3 A0A1Y1JZI9 A0A2C9KLV8 A0A2A3EEU2 A0A3M1M8S6 A0A3P9JBI1 A0A224XWA0 C1BRB9 A0A3P9HVP1 A0A1X7UCT0 A0A069DVY8 A0A146MVE7 A0A3Q3EIB4 A0A146W0H0 A0A0V0GBM8 A0A3B3TSC8 A0A3M1MHJ1 A0A3B3IFE4 C1BQE7 A0A0P4VJC4 R4G3R0 A0A2K6F0A1 A0A336M697 A0A2I4CG74 C3KJM3 H0WIN4 A0A085MAB3 A0A1W5AY64 A0A146ZXA2 A0A0S7HFE7 A0A3B3XP86 A0A3B5MTJ1 A0A3B4BGB0 E0VAS8 A0A226E348 A0A131Z494 G3MSD3 A0A067QY57 A0A3B5AHI7 C3KH08 A0A3B4TWR5 A0A3B3D813 A0A131XL15 A0A3Q2DIC4 A0A3B1KHR4 A0A1A7XWU2 A0A3B4F597 A0A3P9DIR8 A0A3P8SEV9 G5B8Y4 A0A3Q4GIG3 I3MHC8 I3KHW6 A0A1H4EH84 A0A3B5QXT5 G7PHN9 A0A2K6NJW1 A0A0M4EAK3 H2TRV4 A0A2K6JSX5 A0A3Q1JRQ1 A0A0B6YBB3 A0A3Q1B131 T1F395 A0A3S1B6V3 A0A2Y9JA97
PDB
1ONI
E-value=8.76465e-27,
Score=294
Ontologies
PANTHER
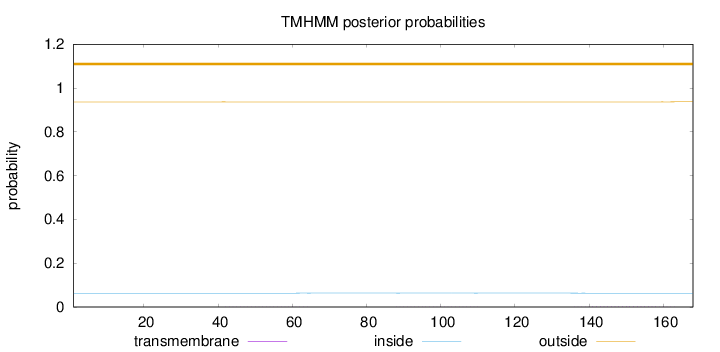
Topology
Length:
168
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04307
Exp number, first 60 AAs:
0.00617
Total prob of N-in:
0.06336
outside
1 - 168
Population Genetic Test Statistics
Pi
237.197353
Theta
188.019373
Tajima's D
0.828402
CLR
0
CSRT
0.610569471526424
Interpretation
Uncertain
