Pre Gene Modal
BGIBMGA011783
Annotation
autophagy_related_protein_Atg8_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.668 Nuclear Reliability : 2.103
Sequence
CDS
ATGAAATTCCAATACAAAGAAGAACATTCATTTGAGAAGAGAAAGGCTGAGGGCGAAAAAATCCGCAGGAAATATCCAGATCGCGTTCCTGTAATTGTAGAGAAAGCGCCGAAGGCTAGGCTTGGAGACCTCGACAAAAAGAAGTATTTAGTTCCGTCTGATTTAACAGTTGGCCAGTTCTACTTCTTGATCCGGAAACGTATTCACCTGCGGCCTGAAGACGCACTTTTCTTCTTCGTGAACAATGTCATTCCACCCACATCTGCAACAATGGGGTCTCTCTACCAAGAACACCATGATGAAGATTTCTTTCTATACATAGCATTTTCCGACGAAAATGTCTATGGAAATTAA
Protein
MKFQYKEEHSFEKRKAEGEKIRRKYPDRVPVIVEKAPKARLGDLDKKKYLVPSDLTVGQFYFLIRKRIHLRPEDALFFFVNNVIPPTSATMGSLYQEHHDEDFFLYIAFSDENVYGN
Summary
Similarity
Belongs to the ATG8 family.
Uniprot
Q2F5R5
A0A0L7LIF5
J7G5G3
A0A1Z2TIV3
I3W7D5
A0A194QP92
+ More
S4PX79 S4U811 S4U3I5 I4DJ07 A0A212FDC5 A0A2J7RF63 X5I348 A0A2P8XE72 R4ULD6 A0A1B6C2W9 T1E2N0 A0A1B6GSU5 A0A2H4NI26 Q1W292 D2A3P4 B4N2G9 A0A3B0JF84 B3MSC0 Q9W2S2 A0A023EDG5 B4L2S7 B3NW04 B4R7M5 B4PXZ4 B4IDT6 A0A3G4S8N6 A0A3S3QPM4 A0A1Q3FJN6 B0W6U4 B4MER6 A0A0M4F9U6 B4JK65 A2I460 A0A0E3IQF3 A0A1S3CZM9 B5DLB9 B4H2U2 A0A1W4V2T1 A0A1L8EFI4 C4N190 E0VE87 V5I994 A0A0K8U091 A0A0C9PK21 A0A0A1X8S6 K0BVA5 Q173G7 A0A0P5CZH7 K1PXH7 E2C3G0 A0A0P5C5R6 A0A164PTL5 A0A084WR93 A0A346QR78 W8CBZ1 A0A1Y1M121 A0A336LXS2 Q5MM86 A0A1J1IGW5 A0A0P5RS48 A0A0P5X341 A0A0P5Y131 A0A0L0BZZ9 A0A1A9X7Z3 A0A1B0AJZ0 A0A0L8ICY3 A0A182Y428 D3TLZ5 A0A0P4ZTK1 A0A232FAL2 A0A2C9K2E5 D7RP03 H9XQ24 A0A182VZW5 V9LVC3 V9LV30 W5JNN1 A0A0A9YNQ6 A0A0P5A0Q8 A0A210PX58 A0A182RTH4 A0A2M4C2T3 A0A182MCB0 A0A2M3YYL1 A0A067QS99 A0A182JNN2 A0A2M4A0L9 V4ADG8 R4WR84 A0A151IIQ3 A0A195DNI1 F4X5H4 A0A195BPV8 A0A0J7KM75
S4PX79 S4U811 S4U3I5 I4DJ07 A0A212FDC5 A0A2J7RF63 X5I348 A0A2P8XE72 R4ULD6 A0A1B6C2W9 T1E2N0 A0A1B6GSU5 A0A2H4NI26 Q1W292 D2A3P4 B4N2G9 A0A3B0JF84 B3MSC0 Q9W2S2 A0A023EDG5 B4L2S7 B3NW04 B4R7M5 B4PXZ4 B4IDT6 A0A3G4S8N6 A0A3S3QPM4 A0A1Q3FJN6 B0W6U4 B4MER6 A0A0M4F9U6 B4JK65 A2I460 A0A0E3IQF3 A0A1S3CZM9 B5DLB9 B4H2U2 A0A1W4V2T1 A0A1L8EFI4 C4N190 E0VE87 V5I994 A0A0K8U091 A0A0C9PK21 A0A0A1X8S6 K0BVA5 Q173G7 A0A0P5CZH7 K1PXH7 E2C3G0 A0A0P5C5R6 A0A164PTL5 A0A084WR93 A0A346QR78 W8CBZ1 A0A1Y1M121 A0A336LXS2 Q5MM86 A0A1J1IGW5 A0A0P5RS48 A0A0P5X341 A0A0P5Y131 A0A0L0BZZ9 A0A1A9X7Z3 A0A1B0AJZ0 A0A0L8ICY3 A0A182Y428 D3TLZ5 A0A0P4ZTK1 A0A232FAL2 A0A2C9K2E5 D7RP03 H9XQ24 A0A182VZW5 V9LVC3 V9LV30 W5JNN1 A0A0A9YNQ6 A0A0P5A0Q8 A0A210PX58 A0A182RTH4 A0A2M4C2T3 A0A182MCB0 A0A2M3YYL1 A0A067QS99 A0A182JNN2 A0A2M4A0L9 V4ADG8 R4WR84 A0A151IIQ3 A0A195DNI1 F4X5H4 A0A195BPV8 A0A0J7KM75
Pubmed
19121390
19470186
20606273
26227816
22830988
28595926
+ More
22629397 26354079 23622113 22651552 22118469 29403074 24330624 29125860 18362917 19820115 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 17550304 25727880 15632085 19576987 20566863 25830018 25348373 17510324 22992520 20798317 24438588 24495485 28004739 26108605 25244985 20353571 28648823 15562597 22771962 20920257 23761445 25401762 28812685 24845553 23254933 23691247 21719571
22629397 26354079 23622113 22651552 22118469 29403074 24330624 29125860 18362917 19820115 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 17550304 25727880 15632085 19576987 20566863 25830018 25348373 17510324 22992520 20798317 24438588 24495485 28004739 26108605 25244985 20353571 28648823 15562597 22771962 20920257 23761445 25401762 28812685 24845553 23254933 23691247 21719571
EMBL
BABH01012002
DQ311358
FJ416330
FJ595955
ABD36302.1
ACJ46064.1
+ More
ACO53446.1 JTDY01001049 KOB75016.1 JN705946 AFP66874.1 MF069154 ODYU01003871 ASA69721.1 SOQ43173.1 JQ739159 AFK91514.1 KQ461190 KPJ07169.1 GAIX01005508 JAA87052.1 JX183217 AGH68882.1 JX183216 AGH68881.1 AK401275 KQ459590 BAM17897.1 KPI97374.1 AGBW02009074 OWR51741.1 NEVH01004413 PNF39469.1 AB856588 BAO58963.1 PYGN01002564 PSN30272.1 KC741165 AGM32989.1 GEDC01029689 JAS07609.1 GALA01000554 JAA94298.1 GECZ01024772 GECZ01004277 JAS44997.1 JAS65492.1 MF038045 ATV91621.1 DQ445535 GEBQ01017747 ABD98773.1 JAT22230.1 KQ971348 EFA05804.2 CH963925 EDW78558.1 OUUW01000003 SPP78872.1 CH902622 EDV34675.1 AE014298 AY122152 AAF46617.1 AAM52664.1 JXUM01098676 JXUM01098677 JXUM01098678 GAPW01006161 GAPW01006160 KQ564439 MH243747 JAC07437.1 KXJ72272.1 QBI60017.1 CH933810 EDW06894.1 CH954180 EDV46137.1 CM000366 EDX17584.1 CM000162 EDX02966.1 CH480830 EDW45744.1 MG948471 AYU75107.1 NCKU01001520 NCKU01001477 NCKU01001475 NCKU01000421 RWS11926.1 RWS12022.1 RWS12025.1 RWS15566.1 GFDL01007311 JAV27734.1 DS231849 EDS36925.1 CH940664 EDW63041.1 CP012528 ALC49070.1 CH916370 EDV99967.1 EF070563 ABM55629.1 KM676434 AJE26300.1 CH379063 EDY72318.1 CH479204 EDW30659.1 GFDG01001460 JAV17339.1 EZ048863 ACN69155.1 DS235088 EEB11693.1 GALX01003330 JAB65136.1 GDHF01032368 JAI19946.1 GBYB01001343 JAG71110.1 GBXI01013843 GBXI01006588 JAD00449.1 JAD07704.1 JQ796071 GAKP01009794 AFS89701.1 JAC49158.1 CH477425 EAT41168.1 GDIP01179406 GDIQ01062927 JAJ43996.1 JAN31810.1 JH817066 EKC26388.1 GL452320 EFN77498.1 GDIP01179405 JAJ43997.1 LRGB01002568 KZS07129.1 ATLV01026002 KE525404 KFB52737.1 MG910480 AXR98468.1 GAMC01004366 JAC02190.1 GEZM01044969 JAV78005.1 UFQS01000289 UFQT01000289 SSX02478.1 SSX22852.1 AY736002 AAW21996.1 CVRI01000047 CRK97673.1 GDIQ01102613 JAL49113.1 GDIP01078565 JAM25150.1 GDIP01066122 JAM37593.1 JRES01001093 KNC25630.1 KQ415974 KOF99368.1 CCAG010016762 EZ422447 ADD18723.1 GDIP01208751 JAJ14651.1 NNAY01000520 OXU27884.1 HM107004 ADI56518.1 JN874925 JN874926 AFH01335.1 JQ410230 AFV99179.1 JQ410231 AFV99180.1 ADMH02000547 ETN65982.1 GBHO01010333 JAG33271.1 GDIP01208750 JAJ14652.1 NEDP02005424 OWF41054.1 GGFJ01010463 MBW59604.1 AXCM01000275 GGFM01000608 MBW21359.1 KK853682 KDQ97907.1 GGFK01000974 MBW34295.1 KB202283 ESO91351.1 AK417177 BAN20392.1 KQ977459 KYN02565.1 KQ980713 KYN14387.1 GL888716 EGI58330.1 KQ976432 KYM87529.1 LBMM01005645 KMQ91336.1
ACO53446.1 JTDY01001049 KOB75016.1 JN705946 AFP66874.1 MF069154 ODYU01003871 ASA69721.1 SOQ43173.1 JQ739159 AFK91514.1 KQ461190 KPJ07169.1 GAIX01005508 JAA87052.1 JX183217 AGH68882.1 JX183216 AGH68881.1 AK401275 KQ459590 BAM17897.1 KPI97374.1 AGBW02009074 OWR51741.1 NEVH01004413 PNF39469.1 AB856588 BAO58963.1 PYGN01002564 PSN30272.1 KC741165 AGM32989.1 GEDC01029689 JAS07609.1 GALA01000554 JAA94298.1 GECZ01024772 GECZ01004277 JAS44997.1 JAS65492.1 MF038045 ATV91621.1 DQ445535 GEBQ01017747 ABD98773.1 JAT22230.1 KQ971348 EFA05804.2 CH963925 EDW78558.1 OUUW01000003 SPP78872.1 CH902622 EDV34675.1 AE014298 AY122152 AAF46617.1 AAM52664.1 JXUM01098676 JXUM01098677 JXUM01098678 GAPW01006161 GAPW01006160 KQ564439 MH243747 JAC07437.1 KXJ72272.1 QBI60017.1 CH933810 EDW06894.1 CH954180 EDV46137.1 CM000366 EDX17584.1 CM000162 EDX02966.1 CH480830 EDW45744.1 MG948471 AYU75107.1 NCKU01001520 NCKU01001477 NCKU01001475 NCKU01000421 RWS11926.1 RWS12022.1 RWS12025.1 RWS15566.1 GFDL01007311 JAV27734.1 DS231849 EDS36925.1 CH940664 EDW63041.1 CP012528 ALC49070.1 CH916370 EDV99967.1 EF070563 ABM55629.1 KM676434 AJE26300.1 CH379063 EDY72318.1 CH479204 EDW30659.1 GFDG01001460 JAV17339.1 EZ048863 ACN69155.1 DS235088 EEB11693.1 GALX01003330 JAB65136.1 GDHF01032368 JAI19946.1 GBYB01001343 JAG71110.1 GBXI01013843 GBXI01006588 JAD00449.1 JAD07704.1 JQ796071 GAKP01009794 AFS89701.1 JAC49158.1 CH477425 EAT41168.1 GDIP01179406 GDIQ01062927 JAJ43996.1 JAN31810.1 JH817066 EKC26388.1 GL452320 EFN77498.1 GDIP01179405 JAJ43997.1 LRGB01002568 KZS07129.1 ATLV01026002 KE525404 KFB52737.1 MG910480 AXR98468.1 GAMC01004366 JAC02190.1 GEZM01044969 JAV78005.1 UFQS01000289 UFQT01000289 SSX02478.1 SSX22852.1 AY736002 AAW21996.1 CVRI01000047 CRK97673.1 GDIQ01102613 JAL49113.1 GDIP01078565 JAM25150.1 GDIP01066122 JAM37593.1 JRES01001093 KNC25630.1 KQ415974 KOF99368.1 CCAG010016762 EZ422447 ADD18723.1 GDIP01208751 JAJ14651.1 NNAY01000520 OXU27884.1 HM107004 ADI56518.1 JN874925 JN874926 AFH01335.1 JQ410230 AFV99179.1 JQ410231 AFV99180.1 ADMH02000547 ETN65982.1 GBHO01010333 JAG33271.1 GDIP01208750 JAJ14652.1 NEDP02005424 OWF41054.1 GGFJ01010463 MBW59604.1 AXCM01000275 GGFM01000608 MBW21359.1 KK853682 KDQ97907.1 GGFK01000974 MBW34295.1 KB202283 ESO91351.1 AK417177 BAN20392.1 KQ977459 KYN02565.1 KQ980713 KYN14387.1 GL888716 EGI58330.1 KQ976432 KYM87529.1 LBMM01005645 KMQ91336.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000235965
+ More
UP000245037 UP000007266 UP000007798 UP000268350 UP000007801 UP000000803 UP000069940 UP000249989 UP000009192 UP000008711 UP000000304 UP000002282 UP000001292 UP000285301 UP000002320 UP000008792 UP000092553 UP000001070 UP000079169 UP000001819 UP000008744 UP000192221 UP000095300 UP000009046 UP000008820 UP000005408 UP000008237 UP000076858 UP000030765 UP000183832 UP000037069 UP000092443 UP000092445 UP000053454 UP000076408 UP000092444 UP000215335 UP000076420 UP000075920 UP000000673 UP000242188 UP000075900 UP000075883 UP000027135 UP000075881 UP000030746 UP000078542 UP000078492 UP000007755 UP000078540 UP000036403
UP000245037 UP000007266 UP000007798 UP000268350 UP000007801 UP000000803 UP000069940 UP000249989 UP000009192 UP000008711 UP000000304 UP000002282 UP000001292 UP000285301 UP000002320 UP000008792 UP000092553 UP000001070 UP000079169 UP000001819 UP000008744 UP000192221 UP000095300 UP000009046 UP000008820 UP000005408 UP000008237 UP000076858 UP000030765 UP000183832 UP000037069 UP000092443 UP000092445 UP000053454 UP000076408 UP000092444 UP000215335 UP000076420 UP000075920 UP000000673 UP000242188 UP000075900 UP000075883 UP000027135 UP000075881 UP000030746 UP000078542 UP000078492 UP000007755 UP000078540 UP000036403
Pfam
PF02991 Atg8
SUPFAM
SSF54236
SSF54236
CDD
ProteinModelPortal
Q2F5R5
A0A0L7LIF5
J7G5G3
A0A1Z2TIV3
I3W7D5
A0A194QP92
+ More
S4PX79 S4U811 S4U3I5 I4DJ07 A0A212FDC5 A0A2J7RF63 X5I348 A0A2P8XE72 R4ULD6 A0A1B6C2W9 T1E2N0 A0A1B6GSU5 A0A2H4NI26 Q1W292 D2A3P4 B4N2G9 A0A3B0JF84 B3MSC0 Q9W2S2 A0A023EDG5 B4L2S7 B3NW04 B4R7M5 B4PXZ4 B4IDT6 A0A3G4S8N6 A0A3S3QPM4 A0A1Q3FJN6 B0W6U4 B4MER6 A0A0M4F9U6 B4JK65 A2I460 A0A0E3IQF3 A0A1S3CZM9 B5DLB9 B4H2U2 A0A1W4V2T1 A0A1L8EFI4 C4N190 E0VE87 V5I994 A0A0K8U091 A0A0C9PK21 A0A0A1X8S6 K0BVA5 Q173G7 A0A0P5CZH7 K1PXH7 E2C3G0 A0A0P5C5R6 A0A164PTL5 A0A084WR93 A0A346QR78 W8CBZ1 A0A1Y1M121 A0A336LXS2 Q5MM86 A0A1J1IGW5 A0A0P5RS48 A0A0P5X341 A0A0P5Y131 A0A0L0BZZ9 A0A1A9X7Z3 A0A1B0AJZ0 A0A0L8ICY3 A0A182Y428 D3TLZ5 A0A0P4ZTK1 A0A232FAL2 A0A2C9K2E5 D7RP03 H9XQ24 A0A182VZW5 V9LVC3 V9LV30 W5JNN1 A0A0A9YNQ6 A0A0P5A0Q8 A0A210PX58 A0A182RTH4 A0A2M4C2T3 A0A182MCB0 A0A2M3YYL1 A0A067QS99 A0A182JNN2 A0A2M4A0L9 V4ADG8 R4WR84 A0A151IIQ3 A0A195DNI1 F4X5H4 A0A195BPV8 A0A0J7KM75
S4PX79 S4U811 S4U3I5 I4DJ07 A0A212FDC5 A0A2J7RF63 X5I348 A0A2P8XE72 R4ULD6 A0A1B6C2W9 T1E2N0 A0A1B6GSU5 A0A2H4NI26 Q1W292 D2A3P4 B4N2G9 A0A3B0JF84 B3MSC0 Q9W2S2 A0A023EDG5 B4L2S7 B3NW04 B4R7M5 B4PXZ4 B4IDT6 A0A3G4S8N6 A0A3S3QPM4 A0A1Q3FJN6 B0W6U4 B4MER6 A0A0M4F9U6 B4JK65 A2I460 A0A0E3IQF3 A0A1S3CZM9 B5DLB9 B4H2U2 A0A1W4V2T1 A0A1L8EFI4 C4N190 E0VE87 V5I994 A0A0K8U091 A0A0C9PK21 A0A0A1X8S6 K0BVA5 Q173G7 A0A0P5CZH7 K1PXH7 E2C3G0 A0A0P5C5R6 A0A164PTL5 A0A084WR93 A0A346QR78 W8CBZ1 A0A1Y1M121 A0A336LXS2 Q5MM86 A0A1J1IGW5 A0A0P5RS48 A0A0P5X341 A0A0P5Y131 A0A0L0BZZ9 A0A1A9X7Z3 A0A1B0AJZ0 A0A0L8ICY3 A0A182Y428 D3TLZ5 A0A0P4ZTK1 A0A232FAL2 A0A2C9K2E5 D7RP03 H9XQ24 A0A182VZW5 V9LVC3 V9LV30 W5JNN1 A0A0A9YNQ6 A0A0P5A0Q8 A0A210PX58 A0A182RTH4 A0A2M4C2T3 A0A182MCB0 A0A2M3YYL1 A0A067QS99 A0A182JNN2 A0A2M4A0L9 V4ADG8 R4WR84 A0A151IIQ3 A0A195DNI1 F4X5H4 A0A195BPV8 A0A0J7KM75
PDB
3M95
E-value=1.22839e-57,
Score=558
Ontologies
KEGG
PATHWAY
GO
PANTHER
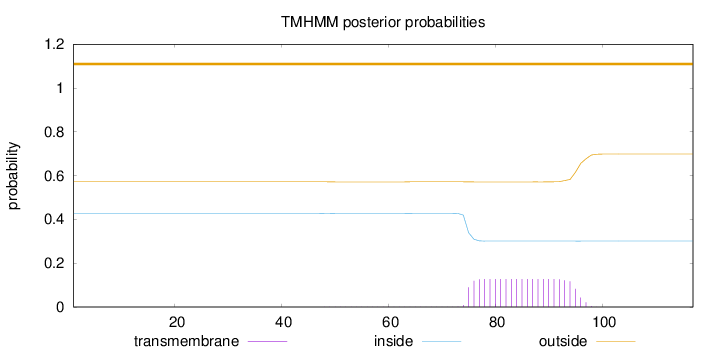
Topology
Subcellular location
Cytoplasm
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.67193
Exp number, first 60 AAs:
0.01219
Total prob of N-in:
0.42787
outside
1 - 117
Population Genetic Test Statistics
Pi
195.987792
Theta
179.253829
Tajima's D
0.293712
CLR
116.7585
CSRT
0.450527473626319
Interpretation
Uncertain
