Gene
KWMTBOMO06626
Annotation
PREDICTED:_U6_snRNA_phosphodiesterase_[Plutella_xylostella]
Full name
U6 snRNA phosphodiesterase
Location in the cell
Nuclear Reliability : 3.617
Sequence
CDS
ATGTCTTGTCTGTCGTATATCAACGATTACGGATCTGATTCAACGAATAGTGACAGTGAAACTGAACAACATCTGTACAAAAGAGTGAAATTACCAGTTCCAGATCTCAGTCGTGTGCGAATAGTTTTAACTGAAGAAACACACATAGACGATCATCAAAAACATAAAGGCCGAAAACGTTCTTTTCCACATGTAAGAGGAAACTGGGCTACTTTCGTCTATGTGCAATATCCTAATGATGACTTTGTTAATAAATTGAGTAGAAAAGTATTAGATGCCTTAACAGAATCATCTCATACTTGGCATAAATGTGAAAATCTCCACATTACTTTATCAAAAACAGTTGTCCTTAATTATCATTGGATAGCACCATTTCACAATTCTTTAAAAAAAACTTTAACAAATATTAAATGTTTTGAACTTGAATTTGATGAGCTGCAAGTATTGTGCAATGAAGATAAATCTAGGACATTTGTTGTGTTGAAAACAAATCGTTTTAGTCATAAGCACTTTTTTGCTATAACTAAGAAGGTTGATGAAATATTGAGTGAATTTAAACTACCAAAATTTTATGATGAACCTTCTTTCCACATGAGCATACTTTGGACTAATGGGGATAAAGAGACAGAGACTAATGGTGCTCTTGACAAATTAAATCAAATCTTGACACCAGATTTTCAGGAATTAGCAAAAAATATTTTTATTGATAAAGTTTACTGTAAAAGCGGAAATAAGACATTTCAGTACCATTTACCATCAGATCGATAG
Protein
MSCLSYINDYGSDSTNSDSETEQHLYKRVKLPVPDLSRVRIVLTEETHIDDHQKHKGRKRSFPHVRGNWATFVYVQYPNDDFVNKLSRKVLDALTESSHTWHKCENLHITLSKTVVLNYHWIAPFHNSLKKTLTNIKCFELEFDELQVLCNEDKSRTFVVLKTNRFSHKHFFAITKKVDEILSEFKLPKFYDEPSFHMSILWTNGDKETETNGALDKLNQILTPDFQELAKNIFIDKVYCKSGNKTFQYHLPSDR
Summary
Description
Phosphodiesterase responsible for the U6 snRNA 3' end processing. Acts as an exoribonuclease (RNase) responsible for trimming the poly(U) tract of the last nucleotides in the pre-U6 snRNA molecule, leading to the formation of mature U6 snRNA.
Phosphodiesterase responsible for the U6 snRNA 3' end processing. Acts as an exoribonuclease (RNase) responsible for trimming the poly(U) tract of the last nucleotides in the pre-U6 snRNA molecule, leading to the formation of mature U6 snRNA 3' end-terminated with a 2',3'-cyclic phosphate.
Phosphodiesterase responsible for the U6 snRNA 3' end processing. Acts as an exoribonuclease (RNase) responsible for trimming the poly(U) tract of the last nucleotides in the pre-U6 snRNA molecule, leading to the formation of mature U6 snRNA 3' end-terminated with a 2',3'-cyclic phosphate.
Cofactor
Zn(2+)
Similarity
Belongs to the 2H phosphoesterase superfamily. USB1 family.
Belongs to the peptidase M10A family.
Belongs to the peptidase M10A family.
Keywords
Complete proteome
Hydrolase
Nuclease
Nucleus
Reference proteome
3D-structure
Alternative splicing
Polymorphism
Feature
chain U6 snRNA phosphodiesterase
splice variant In isoform 3.
sequence variant In dbSNP:rs35025252.
splice variant In isoform 3.
sequence variant In dbSNP:rs35025252.
Uniprot
A0A2H1VP25
A0A194PVL7
A0A194QNT5
A0A212F5X0
A0A2A4JLF2
A0A3S2P4V3
+ More
A0A1Y1LAA8 A0A1W4WWV0 L8IE31 W5NSK8 Q0II50 F7E331 A0A146LR70 A0A2K6DT55 G7Q194 A0A0A9W5M9 A0A0K8T016 A0A2K5LU96 A0A2K5WCV2 A0A2K5Y0R0 A0A096NIX9 G7NPV9 G1P970 A0A1U7QTA9 A0A2K6NZ53 A0A1S2ZJ59 A0A3Q7VEU7 A0A1U7U1T1 A0A151MSJ5 A0A2J8VZG7 F7F9N5 A0A0D9QXM2 A0A2I2YWK7 A0A340Y540 Q91W78 A0A0R4J0E0 M3VYZ4 A0A151MSJ7 K7BU00 A0A2R9CAG8 F6TF64 A0A383YXP5 A0A341BNB4 A0A2Y9LPJ2 L5KXC0 A0A384D2F4 A0A2K6U1Q8 A0A2Y9ERW4 F6TSW3 A0A2K5PYS5 A0A2Y9DBP5 A0A2U3VLG3 A0A2U4BDZ5 A0A1U7RLK3 A0A2K6G4P4 A0A2K5JF53 A0A1I8Q4D6 A0A2K5ESR1 A0A2K5JF38 A0A0K8WIV6 A0A024R6V6 Q9BQ65 W8BAF1 A0A2D0RGM5 A0A2D0RGL2 Q5I0I5 A0A2D0RGM1 A0A3Q7QH67 A0A182QX31 K9IZD0 A0A2K6N312 D2GTY3 W5LWP7 A0A3Q7RW66 B4JFE2 A0A1S3EWS7 A0A250XXB1 G1MFP5 G3I3J6 W5LWK0 A0A3Q7SVC9 A0A0K8V4Q3 A0A034WGQ5 L9KU30 A0A2J8VZJ9 A0A3B1JHC3 H2QB80 A0A2U3YQ15 A0A2J8MVQ0 A0A1I8M3B6 L5MIK6 A0A2J8VZM2 A0A3Q0S5S4 F1RFS6 S9X8T8 V9L8C2 U6DCX2 A0A2Y9KTD7 A0A182VRX7 A0A212DC28
A0A1Y1LAA8 A0A1W4WWV0 L8IE31 W5NSK8 Q0II50 F7E331 A0A146LR70 A0A2K6DT55 G7Q194 A0A0A9W5M9 A0A0K8T016 A0A2K5LU96 A0A2K5WCV2 A0A2K5Y0R0 A0A096NIX9 G7NPV9 G1P970 A0A1U7QTA9 A0A2K6NZ53 A0A1S2ZJ59 A0A3Q7VEU7 A0A1U7U1T1 A0A151MSJ5 A0A2J8VZG7 F7F9N5 A0A0D9QXM2 A0A2I2YWK7 A0A340Y540 Q91W78 A0A0R4J0E0 M3VYZ4 A0A151MSJ7 K7BU00 A0A2R9CAG8 F6TF64 A0A383YXP5 A0A341BNB4 A0A2Y9LPJ2 L5KXC0 A0A384D2F4 A0A2K6U1Q8 A0A2Y9ERW4 F6TSW3 A0A2K5PYS5 A0A2Y9DBP5 A0A2U3VLG3 A0A2U4BDZ5 A0A1U7RLK3 A0A2K6G4P4 A0A2K5JF53 A0A1I8Q4D6 A0A2K5ESR1 A0A2K5JF38 A0A0K8WIV6 A0A024R6V6 Q9BQ65 W8BAF1 A0A2D0RGM5 A0A2D0RGL2 Q5I0I5 A0A2D0RGM1 A0A3Q7QH67 A0A182QX31 K9IZD0 A0A2K6N312 D2GTY3 W5LWP7 A0A3Q7RW66 B4JFE2 A0A1S3EWS7 A0A250XXB1 G1MFP5 G3I3J6 W5LWK0 A0A3Q7SVC9 A0A0K8V4Q3 A0A034WGQ5 L9KU30 A0A2J8VZJ9 A0A3B1JHC3 H2QB80 A0A2U3YQ15 A0A2J8MVQ0 A0A1I8M3B6 L5MIK6 A0A2J8VZM2 A0A3Q0S5S4 F1RFS6 S9X8T8 V9L8C2 U6DCX2 A0A2Y9KTD7 A0A182VRX7 A0A212DC28
EC Number
3.1.4.-
Pubmed
26354079
22118469
28004739
22751099
20809919
19892987
+ More
26823975 22002653 25401762 17431167 25319552 21993624 25362486 22293439 18464734 16141072 15489334 19468303 17975172 22722832 25243066 23258410 17495919 11181995 14702039 15616553 20004881 20503306 22899009 23190533 24495485 20010809 17994087 28087693 21804562 29704459 25348373 23385571 25329095 16136131 25315136 23149746 24402279 29294181
26823975 22002653 25401762 17431167 25319552 21993624 25362486 22293439 18464734 16141072 15489334 19468303 17975172 22722832 25243066 23258410 17495919 11181995 14702039 15616553 20004881 20503306 22899009 23190533 24495485 20010809 17994087 28087693 21804562 29704459 25348373 23385571 25329095 16136131 25315136 23149746 24402279 29294181
EMBL
ODYU01003588
SOQ42547.1
KQ459590
KPI97367.1
KQ461190
KPJ07177.1
+ More
AGBW02010125 OWR49123.1 NWSH01001177 PCG72242.1 RSAL01000414 RVE41831.1 GEZM01062320 JAV69828.1 JH881589 ELR53412.1 AMGL01027930 BC122807 GDHC01008298 GDHC01007400 JAQ10331.1 JAQ11229.1 CM001295 EHH60432.1 GBHO01043454 JAG00150.1 GBRD01006957 JAG58864.1 AQIA01043035 AHZZ02018797 AHZZ02018798 JSUE03026294 JSUE03026295 JU336397 JU472843 JV045352 CM001272 AFE80150.1 AFH29647.1 AFI35423.1 EHH31711.1 AAPE02069114 AKHW03005169 KYO27487.1 NDHI03003406 PNJ62932.1 AQIB01137321 CABD030101248 AK135527 AK160901 AK168683 BC016418 AC165414 AANG04002512 KYO27486.1 GABC01010570 GABF01003301 GABD01005551 GABE01011003 NBAG03000242 JAA00768.1 JAA18844.1 JAA27549.1 JAA33736.1 PNI63608.1 AJFE02083779 AJFE02083780 AJFE02083781 GAMT01009349 GAMS01010386 GAMR01010776 GAMQ01000183 GAMP01008688 JAB02512.1 JAB12750.1 JAB23156.1 JAB41668.1 JAB44067.1 KB030537 ELK15433.1 GDHF01001218 JAI51096.1 CH471092 EAW82964.1 AK023216 AK301494 AK303121 AC010543 AC012182 BC004415 BC006291 BC007774 BC010099 BC021554 GAMC01012547 JAB94008.1 BC088280 AXCN02002238 GABZ01007461 JAA46064.1 GL192338 EFB22239.1 CH916369 EDV93423.1 GFFV01003939 JAV36006.1 ACTA01016790 JH001191 RAZU01000121 EGW12718.1 RLQ71457.1 GDHF01032308 GDHF01018729 JAI20006.1 JAI33585.1 GAKP01005638 GAKP01005637 GAKP01005636 JAC53315.1 KB320660 ELW66208.1 PNJ62928.1 AACZ04057490 PNI63610.1 PNI63607.1 KB098932 ELK38464.1 PNJ62933.1 AEMK02000040 KB016731 EPY84773.1 JW875551 AFP08068.1 HAAF01008143 CCP79967.1 MKHE01000004 OWK15776.1
AGBW02010125 OWR49123.1 NWSH01001177 PCG72242.1 RSAL01000414 RVE41831.1 GEZM01062320 JAV69828.1 JH881589 ELR53412.1 AMGL01027930 BC122807 GDHC01008298 GDHC01007400 JAQ10331.1 JAQ11229.1 CM001295 EHH60432.1 GBHO01043454 JAG00150.1 GBRD01006957 JAG58864.1 AQIA01043035 AHZZ02018797 AHZZ02018798 JSUE03026294 JSUE03026295 JU336397 JU472843 JV045352 CM001272 AFE80150.1 AFH29647.1 AFI35423.1 EHH31711.1 AAPE02069114 AKHW03005169 KYO27487.1 NDHI03003406 PNJ62932.1 AQIB01137321 CABD030101248 AK135527 AK160901 AK168683 BC016418 AC165414 AANG04002512 KYO27486.1 GABC01010570 GABF01003301 GABD01005551 GABE01011003 NBAG03000242 JAA00768.1 JAA18844.1 JAA27549.1 JAA33736.1 PNI63608.1 AJFE02083779 AJFE02083780 AJFE02083781 GAMT01009349 GAMS01010386 GAMR01010776 GAMQ01000183 GAMP01008688 JAB02512.1 JAB12750.1 JAB23156.1 JAB41668.1 JAB44067.1 KB030537 ELK15433.1 GDHF01001218 JAI51096.1 CH471092 EAW82964.1 AK023216 AK301494 AK303121 AC010543 AC012182 BC004415 BC006291 BC007774 BC010099 BC021554 GAMC01012547 JAB94008.1 BC088280 AXCN02002238 GABZ01007461 JAA46064.1 GL192338 EFB22239.1 CH916369 EDV93423.1 GFFV01003939 JAV36006.1 ACTA01016790 JH001191 RAZU01000121 EGW12718.1 RLQ71457.1 GDHF01032308 GDHF01018729 JAI20006.1 JAI33585.1 GAKP01005638 GAKP01005637 GAKP01005636 JAC53315.1 KB320660 ELW66208.1 PNJ62928.1 AACZ04057490 PNI63610.1 PNI63607.1 KB098932 ELK38464.1 PNJ62933.1 AEMK02000040 KB016731 EPY84773.1 JW875551 AFP08068.1 HAAF01008143 CCP79967.1 MKHE01000004 OWK15776.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000218220
UP000283053
UP000192223
+ More
UP000002356 UP000009136 UP000002281 UP000233120 UP000009130 UP000233060 UP000233100 UP000233140 UP000028761 UP000006718 UP000001074 UP000189706 UP000233200 UP000079721 UP000286642 UP000189704 UP000050525 UP000002279 UP000029965 UP000001519 UP000265300 UP000000589 UP000011712 UP000240080 UP000008225 UP000261681 UP000252040 UP000248483 UP000010552 UP000261680 UP000233220 UP000248484 UP000002280 UP000233040 UP000248480 UP000245340 UP000245320 UP000189705 UP000233160 UP000233080 UP000095300 UP000233020 UP000005640 UP000221080 UP000002494 UP000286641 UP000075886 UP000233180 UP000018468 UP000001070 UP000081671 UP000008912 UP000001075 UP000273346 UP000286640 UP000011518 UP000018467 UP000002277 UP000245341 UP000095301 UP000261340 UP000008227 UP000248482 UP000075920
UP000002356 UP000009136 UP000002281 UP000233120 UP000009130 UP000233060 UP000233100 UP000233140 UP000028761 UP000006718 UP000001074 UP000189706 UP000233200 UP000079721 UP000286642 UP000189704 UP000050525 UP000002279 UP000029965 UP000001519 UP000265300 UP000000589 UP000011712 UP000240080 UP000008225 UP000261681 UP000252040 UP000248483 UP000010552 UP000261680 UP000233220 UP000248484 UP000002280 UP000233040 UP000248480 UP000245340 UP000245320 UP000189705 UP000233160 UP000233080 UP000095300 UP000233020 UP000005640 UP000221080 UP000002494 UP000286641 UP000075886 UP000233180 UP000018468 UP000001070 UP000081671 UP000008912 UP000001075 UP000273346 UP000286640 UP000011518 UP000018467 UP000002277 UP000245341 UP000095301 UP000261340 UP000008227 UP000248482 UP000075920
Pfam
Interpro
IPR027521
Usb1
+ More
IPR009097 cNuc_Pdiesterase
IPR000571 Znf_CCCH
IPR028729 MMP15
IPR036365 PGBD-like_sf
IPR006026 Peptidase_Metallo
IPR018487 Hemopexin-like_repeat
IPR000585 Hemopexin-like_dom
IPR021805 Pept_M10A_metallopeptidase_C
IPR024079 MetalloPept_cat_dom_sf
IPR018486 Hemopexin_CS
IPR033739 M10A_MMP
IPR021190 Pept_M10A
IPR036375 Hemopexin-like_dom_sf
IPR021158 Pept_M10A_Zn_BS
IPR001818 Pept_M10_metallopeptidase
IPR002477 Peptidoglycan-bd-like
IPR009097 cNuc_Pdiesterase
IPR000571 Znf_CCCH
IPR028729 MMP15
IPR036365 PGBD-like_sf
IPR006026 Peptidase_Metallo
IPR018487 Hemopexin-like_repeat
IPR000585 Hemopexin-like_dom
IPR021805 Pept_M10A_metallopeptidase_C
IPR024079 MetalloPept_cat_dom_sf
IPR018486 Hemopexin_CS
IPR033739 M10A_MMP
IPR021190 Pept_M10A
IPR036375 Hemopexin-like_dom_sf
IPR021158 Pept_M10A_Zn_BS
IPR001818 Pept_M10_metallopeptidase
IPR002477 Peptidoglycan-bd-like
Gene 3D
ProteinModelPortal
A0A2H1VP25
A0A194PVL7
A0A194QNT5
A0A212F5X0
A0A2A4JLF2
A0A3S2P4V3
+ More
A0A1Y1LAA8 A0A1W4WWV0 L8IE31 W5NSK8 Q0II50 F7E331 A0A146LR70 A0A2K6DT55 G7Q194 A0A0A9W5M9 A0A0K8T016 A0A2K5LU96 A0A2K5WCV2 A0A2K5Y0R0 A0A096NIX9 G7NPV9 G1P970 A0A1U7QTA9 A0A2K6NZ53 A0A1S2ZJ59 A0A3Q7VEU7 A0A1U7U1T1 A0A151MSJ5 A0A2J8VZG7 F7F9N5 A0A0D9QXM2 A0A2I2YWK7 A0A340Y540 Q91W78 A0A0R4J0E0 M3VYZ4 A0A151MSJ7 K7BU00 A0A2R9CAG8 F6TF64 A0A383YXP5 A0A341BNB4 A0A2Y9LPJ2 L5KXC0 A0A384D2F4 A0A2K6U1Q8 A0A2Y9ERW4 F6TSW3 A0A2K5PYS5 A0A2Y9DBP5 A0A2U3VLG3 A0A2U4BDZ5 A0A1U7RLK3 A0A2K6G4P4 A0A2K5JF53 A0A1I8Q4D6 A0A2K5ESR1 A0A2K5JF38 A0A0K8WIV6 A0A024R6V6 Q9BQ65 W8BAF1 A0A2D0RGM5 A0A2D0RGL2 Q5I0I5 A0A2D0RGM1 A0A3Q7QH67 A0A182QX31 K9IZD0 A0A2K6N312 D2GTY3 W5LWP7 A0A3Q7RW66 B4JFE2 A0A1S3EWS7 A0A250XXB1 G1MFP5 G3I3J6 W5LWK0 A0A3Q7SVC9 A0A0K8V4Q3 A0A034WGQ5 L9KU30 A0A2J8VZJ9 A0A3B1JHC3 H2QB80 A0A2U3YQ15 A0A2J8MVQ0 A0A1I8M3B6 L5MIK6 A0A2J8VZM2 A0A3Q0S5S4 F1RFS6 S9X8T8 V9L8C2 U6DCX2 A0A2Y9KTD7 A0A182VRX7 A0A212DC28
A0A1Y1LAA8 A0A1W4WWV0 L8IE31 W5NSK8 Q0II50 F7E331 A0A146LR70 A0A2K6DT55 G7Q194 A0A0A9W5M9 A0A0K8T016 A0A2K5LU96 A0A2K5WCV2 A0A2K5Y0R0 A0A096NIX9 G7NPV9 G1P970 A0A1U7QTA9 A0A2K6NZ53 A0A1S2ZJ59 A0A3Q7VEU7 A0A1U7U1T1 A0A151MSJ5 A0A2J8VZG7 F7F9N5 A0A0D9QXM2 A0A2I2YWK7 A0A340Y540 Q91W78 A0A0R4J0E0 M3VYZ4 A0A151MSJ7 K7BU00 A0A2R9CAG8 F6TF64 A0A383YXP5 A0A341BNB4 A0A2Y9LPJ2 L5KXC0 A0A384D2F4 A0A2K6U1Q8 A0A2Y9ERW4 F6TSW3 A0A2K5PYS5 A0A2Y9DBP5 A0A2U3VLG3 A0A2U4BDZ5 A0A1U7RLK3 A0A2K6G4P4 A0A2K5JF53 A0A1I8Q4D6 A0A2K5ESR1 A0A2K5JF38 A0A0K8WIV6 A0A024R6V6 Q9BQ65 W8BAF1 A0A2D0RGM5 A0A2D0RGL2 Q5I0I5 A0A2D0RGM1 A0A3Q7QH67 A0A182QX31 K9IZD0 A0A2K6N312 D2GTY3 W5LWP7 A0A3Q7RW66 B4JFE2 A0A1S3EWS7 A0A250XXB1 G1MFP5 G3I3J6 W5LWK0 A0A3Q7SVC9 A0A0K8V4Q3 A0A034WGQ5 L9KU30 A0A2J8VZJ9 A0A3B1JHC3 H2QB80 A0A2U3YQ15 A0A2J8MVQ0 A0A1I8M3B6 L5MIK6 A0A2J8VZM2 A0A3Q0S5S4 F1RFS6 S9X8T8 V9L8C2 U6DCX2 A0A2Y9KTD7 A0A182VRX7 A0A212DC28
PDB
4H7W
E-value=3.70175e-28,
Score=308
Ontologies
GO
PANTHER
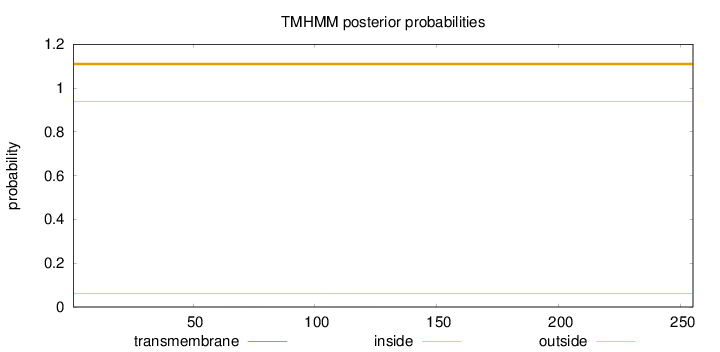
Topology
Subcellular location
Nucleus
Length:
255
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00753
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06175
outside
1 - 255
Population Genetic Test Statistics
Pi
120.223984
Theta
138.957278
Tajima's D
-0.497192
CLR
110.633775
CSRT
0.249787510624469
Interpretation
Uncertain
