Gene
KWMTBOMO06624
Pre Gene Modal
BGIBMGA011691
Annotation
PREDICTED:_glutamate_receptor_ionotropic?_NMDA_2B-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.813
Sequence
CDS
ATGCAGACGCATGCAGCTCTAATGGCGCTGGCTGCGCTGGGAGCGCTAGCGGCGGCGGCGGCGGCGGAGCGCGGCAGTGGCATCAAAGTCGGCAGCGGTGGTGGAATGGCCAGTAGTCGAGATGGTGGCCGTGGAAGTGGTGTACGCATCGGAGACGGAATCAGGCGTCCACGAACTTCACCTGCGCCTCCTCCTCCTCGAGCTCCTTCTTCCATCACCGCTGCTCTCGTCGTGCCACATAAGGCCTTTGGCACACGAGATTACACCAAGGCCGAAAAAGCTGCTCTATCAAAATTACCGCGAAAACTGAAACTTTTTTCTCAAGTGCGCCTGAACATAACGCTCTCTACGCAAGGCTTGACGCCTAGTCCTATGTCTATCCTGGACTCGCTATGTAAAGAATTTCTAGCAGTCAACGTGTCTGCTATCCTTTATCTTATGAATCACGAACAATACGGGCGCTCTACTGCCTCTGCACAATATTTTCTACAACTCGCAGGGTATCTCGGCATACCGGTAATTGCATGGAATGCAGACAACAGCGGGCTTGAGAAGCGCGCGTCACATGCTTCTTTACGTCTTCAATTGGCCCCATCCATTGAACATCAAACAGCCGCGATGCTTTCTATTTTAGAAAGATACAAATGGCATCAGTTCAGCGTCGTCACCTCAGCTATTGCCGGACATGACGATTTCATACAGGCTGTACGTGAAAGAGTGACGGCACTACAAGATCGATTTAAATTCACGATACTTAACGCAATAGTCGTTAAGAGGTCATCAGATCTAAATGAATTGGTCACAAGTGAAGCGCGTGTGATGTTACTGTATGCGACCAGAGAAGAGGCAGCGGAAATACTATCTGCCGCTGGAGACTTACACCTTACCGGCGAAAGTTTTGTGTGGATCGTTACACAAAGTGTATTAGGATCTATGCAACAACCCAACAAATTTCCCGTCGGCATGCTTGGTGTACATTTCGACACGTCGAGTTCTTCTTTGATATCAGAAATAGCAACCGCTGTTAAAGTATTTGCCTACGGCGTGGAGTCGTACATATCCGAGCCTGAGAATGTGCGTTACCCACTGGGCACTAGACTATCGTGCAGTGGCGCGGGAGCAGGCGAAGCACGTTGGTCTACAGGCGAGCGATTCTATAGGCATTTGCGGAATGTCAGCGTTGAAGGAGAAGTAGGGAGACCGAGCATCGAATTCACCCCTGATGGTGAATTACGAGCTGCTGAACTTAAAATTATGAATCTAAGACCTGCCATAGGCGAACAGTTGGTATGGGAAGAAATCGGAACTTGGAACTCTTATCCGAAAGAACGACTCGTCATCAAAGACATTGTTTGGCCTGGCGGTCTACATACTCCACCACAAGGCGTGCCAGAAAAGTTCCACATGCGCATAACCTTCCTAGAAGAACCACCGTATATTAACTTAGCGCCACCGGATCCCGTTAGTGGACGTTGCTCTTTAGACAGAGGTGTGATATGCCGTGTCGCACCGGAAACAGAAGTTGCTGGTCTCGAAGCCGGTACTGCTCACGGAAATAGCTCTCTATACCAGTGTTGCAGTGGATTCTGTATAGATCTGCTGCAACAACTTGCTGAACATCTCGGATTTACCTATGAATTGGTCCGCGTCGAGGATGGACGTTGGGGCACATTGCACTACGGCAAGTGGAACGGTTTGATTGCCGATCTTGTCAACAAGAAAACCGATATGGTTTTAACGTCCTTAATAATAAATTCGGACCGGGAAGCAGTAGTAGATTTCAGTGTGCCATTTATGGAGACCGGTGTTGCAATTGTGGTGGCCAAACGAACAGGCATAATTTCCCCAACGGCTTTCTTAGAACCTTTCGACACTGCTTCGTGGATGCTCGTGGGCGCTGTAGCTATACAAGCTGCTACTTTTTCAATATTCTTCTTCGAATGGATGTCACCGAGTGGATTCGACTGCTCAACTGGCAACTCAAAACGTGTTCCACAAAATCGATTTTCGCTATGTCGCACGTATTGGATTGTTTGGGCAGTTTTATTTCAAGCGTCAGTACACGTTGATTCACCTAGAGGTTTCACAGCTCGTTTCATGACCAACATGTGGGCGATGTTCGCCGTCGTGTTTCTGGCTATATACACAGCTAATTTGGCTGCGTTTATGATAACTCGCGAGGAATACCACGAATTAAGCGGCTTAGACGATCCCAGGATCGCGAGACCACTCTCACAGCGTCCGCCATTAAAATTCGGCACAGTACCGTGGTCACACACAGATGCTACGCTCGCTAAATACTTCCCGGAACCACACGCTTATATGGCTCGGTACAATCGGAGTACAGTTAGCGCAGGAGTGACGAGCGTACTTACGGGAGATCTTGACGCATTCATTTATGACGGTACTGTATTGGATTATCTGGTATCACAAGACGAAGATTGTAGACTGTTGACGGTAGGGGCTTGGTACGCCATGTCCGGGTACGGCCTCGCGTTTACGCGGAACTCGAAATATCTCAGTATGTTCAATAAACGGTTACTAGATTTACGAGCCAATGGTGATCTTGAAAGACTACGAAGGTACTGGATGACTGGAACGTGTAAGCCCAATAAACAAGAGCATAAGTCTTCAGATCCTCTGGCCTTGGAGCAATTCTTATCAGCATTCCTGCTACTTATGGCAGGAATTCTGCTGGCAGCATTGTTGCTCCTGTTAGAACATGTCTATTTTAAATACATGCGCACTCATTTGGCGGCCTCAAAGGCTGGATCGTGTTGTGCGCTAGTTTCGTTATCAATGGGTCAGTCTCTGACTTTTCACGGCGCCGTAGTCGAAGCGGCTGCTCGAGGATTAGGCACAGGCGAACGTGGACACTGTCGCTCCGCCATATGCGCTGCTCAATTATGGCGCGCACGACACGAACGTGATGTAGCCATGGCCAGGGTGAGGCAGCTGGCTGCGGCGTTAGCGGCACACGGGCTGGCGCCGCCGCCACGCCGACTCGCTTCGGCGGCGGCGCTGCTGGCGGGTGGCATGCCGCACGACGCCACGCGGCCGCGCATGCTGCGCCCGCCCGCCGACCTACTGCCGGACCTCGATCGCCCTCTCTCGTGTGGTGATTTGCGATCGAGAGGGCGGACACGAGTAGAAATGGAAACAGTGCTGTGA
Protein
MQTHAALMALAALGALAAAAAAERGSGIKVGSGGGMASSRDGGRGSGVRIGDGIRRPRTSPAPPPPRAPSSITAALVVPHKAFGTRDYTKAEKAALSKLPRKLKLFSQVRLNITLSTQGLTPSPMSILDSLCKEFLAVNVSAILYLMNHEQYGRSTASAQYFLQLAGYLGIPVIAWNADNSGLEKRASHASLRLQLAPSIEHQTAAMLSILERYKWHQFSVVTSAIAGHDDFIQAVRERVTALQDRFKFTILNAIVVKRSSDLNELVTSEARVMLLYATREEAAEILSAAGDLHLTGESFVWIVTQSVLGSMQQPNKFPVGMLGVHFDTSSSSLISEIATAVKVFAYGVESYISEPENVRYPLGTRLSCSGAGAGEARWSTGERFYRHLRNVSVEGEVGRPSIEFTPDGELRAAELKIMNLRPAIGEQLVWEEIGTWNSYPKERLVIKDIVWPGGLHTPPQGVPEKFHMRITFLEEPPYINLAPPDPVSGRCSLDRGVICRVAPETEVAGLEAGTAHGNSSLYQCCSGFCIDLLQQLAEHLGFTYELVRVEDGRWGTLHYGKWNGLIADLVNKKTDMVLTSLIINSDREAVVDFSVPFMETGVAIVVAKRTGIISPTAFLEPFDTASWMLVGAVAIQAATFSIFFFEWMSPSGFDCSTGNSKRVPQNRFSLCRTYWIVWAVLFQASVHVDSPRGFTARFMTNMWAMFAVVFLAIYTANLAAFMITREEYHELSGLDDPRIARPLSQRPPLKFGTVPWSHTDATLAKYFPEPHAYMARYNRSTVSAGVTSVLTGDLDAFIYDGTVLDYLVSQDEDCRLLTVGAWYAMSGYGLAFTRNSKYLSMFNKRLLDLRANGDLERLRRYWMTGTCKPNKQEHKSSDPLALEQFLSAFLLLMAGILLAALLLLLEHVYFKYMRTHLAASKAGSCCALVSLSMGQSLTFHGAVVEAAARGLGTGERGHCRSAICAAQLWRARHERDVAMARVRQLAAALAAHGLAPPPRRLASAAALLAGGMPHDATRPRMLRPPADLLPDLDRPLSCGDLRSRGRTRVEMETVL
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
A0A2H1VNY7
A0A075TD99
H9JQ85
A0A345BF20
A0A212F5V2
A0A194PX73
+ More
A0A0L7LRJ9 A0A2A4JKX7 A0A0F7QHN7 A0A0B5A3G8 A0A0K8TUD8 A0A194QQE4 A0A0V0J153 A0A223HCZ7 A0A139WKM8 A0A0C9RWB2 A0A139WKV8 A0A3L8E2E5 A0A195FTC7 A0A195DKJ2 A0A0L7R9Q3 A0A2A3ENQ0 A0A088A9N9 A0A026WHI8 A0A0L0C858 A0A0M3SBL9 A0A336MC74 A0A1I8M7N9 A0A1I8PVH3 A0A075Q6F7 A0A075Q6G4 A0A075Q6G2 A0A075Q362 A0A0R3P3I3 A0A0M4F7S4 A0A195BG93 A0A0P8XKB0 Q29J15 A0A1S4FIQ8 A0A0Q9XF30 A0A075Q359 B3MRX1 Q16ZR9 B4L761 A0A075QB19 A0A0R3NYN4 B4GTK0 B4PY38 A0A182MIQ3 A0A075Q109 X2JDW0 A0A075Q438 B3P9J4 A0A3B0K250 A0A075Q116 Q9W581 A0A3B0JXG0 A0A182W628 B4I9F7 B4JXG9 A0A3B0JXF2 Q6IWN7 Q16IF5 A0A182RVV6 A0A075Q434 A0A084VQH0 A0A182IWF8 A0A182FMM9 W8C4A0 A0A1J1IN94 K7J2X0 A0A2H8TMD0 A0A1S4G051 A0A1W4VT10 A0A1W4VSC6 A0A1A9X1D8 A0A1B0GFF3 W5JDA1 B0X6V2 A0A151WWC7 E1JJC6 Q7PHP7 A0A1A9YQ94 J9K6V7 B4MB33 A0A195D7B3 B4NPT1 A0A1B0A0B1 Q8MM14 A0A1B0BZ01 A0A2S2QKY9 O76903 B4R2Z6 A0A0C9QFF6 Q8MX77 A0A182GEK7 A0A1L2YXQ5 A0A1L2YXN9
A0A0L7LRJ9 A0A2A4JKX7 A0A0F7QHN7 A0A0B5A3G8 A0A0K8TUD8 A0A194QQE4 A0A0V0J153 A0A223HCZ7 A0A139WKM8 A0A0C9RWB2 A0A139WKV8 A0A3L8E2E5 A0A195FTC7 A0A195DKJ2 A0A0L7R9Q3 A0A2A3ENQ0 A0A088A9N9 A0A026WHI8 A0A0L0C858 A0A0M3SBL9 A0A336MC74 A0A1I8M7N9 A0A1I8PVH3 A0A075Q6F7 A0A075Q6G4 A0A075Q6G2 A0A075Q362 A0A0R3P3I3 A0A0M4F7S4 A0A195BG93 A0A0P8XKB0 Q29J15 A0A1S4FIQ8 A0A0Q9XF30 A0A075Q359 B3MRX1 Q16ZR9 B4L761 A0A075QB19 A0A0R3NYN4 B4GTK0 B4PY38 A0A182MIQ3 A0A075Q109 X2JDW0 A0A075Q438 B3P9J4 A0A3B0K250 A0A075Q116 Q9W581 A0A3B0JXG0 A0A182W628 B4I9F7 B4JXG9 A0A3B0JXF2 Q6IWN7 Q16IF5 A0A182RVV6 A0A075Q434 A0A084VQH0 A0A182IWF8 A0A182FMM9 W8C4A0 A0A1J1IN94 K7J2X0 A0A2H8TMD0 A0A1S4G051 A0A1W4VT10 A0A1W4VSC6 A0A1A9X1D8 A0A1B0GFF3 W5JDA1 B0X6V2 A0A151WWC7 E1JJC6 Q7PHP7 A0A1A9YQ94 J9K6V7 B4MB33 A0A195D7B3 B4NPT1 A0A1B0A0B1 Q8MM14 A0A1B0BZ01 A0A2S2QKY9 O76903 B4R2Z6 A0A0C9QFF6 Q8MX77 A0A182GEK7 A0A1L2YXQ5 A0A1L2YXN9
Pubmed
25027790
19121390
29727827
22118469
26354079
26227816
+ More
25803580 26017144 27006164 28150741 18362917 19820115 30249741 24508170 26108605 26265180 25315136 25657209 15632085 17994087 23185243 17510324 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15823532 26109357 26109356 24438588 24495485 20075255 20920257 23761445 9087549 12364791 14747013 17210077 26483478
25803580 26017144 27006164 28150741 18362917 19820115 30249741 24508170 26108605 26265180 25315136 25657209 15632085 17994087 23185243 17510324 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15823532 26109357 26109356 24438588 24495485 20075255 20920257 23761445 9087549 12364791 14747013 17210077 26483478
EMBL
ODYU01003588
SOQ42545.1
KF768737
AIG51931.1
BABH01012012
MG820673
+ More
AXF48844.1 AGBW02010125 OWR49121.1 KQ459590 KPI97364.1 JTDY01000244 KOB78085.1 NWSH01001177 PCG72244.1 LC017780 BAR64794.1 KJ542735 AJD81619.1 GCVX01000126 JAI18104.1 KQ461190 KPJ07180.1 GDKB01000028 JAP38468.1 KY283581 AST36241.1 KQ971326 KYB28456.1 GBYB01013240 JAG83007.1 KYB28457.1 QOIP01000001 RLU26874.1 KQ981272 KYN43865.1 KQ980765 KYN13357.1 KQ414627 KOC67486.1 KZ288212 PBC32826.1 KK107242 EZA54544.1 JRES01000755 KNC28623.1 KP843222 ALD51358.1 UFQS01002934 UFQT01000673 UFQT01002934 SSX14928.1 SSX26393.1 KJ747199 KJ747200 KJ747202 AIG13069.1 AIG13070.1 AIG13072.1 KJ747210 AIG13080.1 KJ747205 AIG13075.1 KJ747206 AIG13076.1 CH379063 KRT06085.1 CP012528 ALC48300.1 KQ976488 KYM83635.1 CH902622 KPU75185.1 EAL32487.3 KRT06083.1 CH933812 KRG07184.1 KJ747201 AIG13071.1 EDV34526.1 CH477485 EAT40152.1 EDW06207.2 KJ747207 AIG13077.1 KRT06084.1 CH479190 EDW25870.1 CM000162 EDX00911.1 AXCM01010787 AXCM01010788 KJ747204 AIG13074.1 AE014298 AHN59259.1 KJ747208 AIG13078.1 CH954183 EDV45490.1 OUUW01000011 SPP86742.1 KJ747209 AIG13079.1 AY616148 AAF45640.2 AAT40461.1 SPP86744.1 CH480825 EDW43838.1 CH916376 EDV95445.1 SPP86745.1 AY616149 AAT40462.1 ACZ95174.1 CH478081 EAT34043.1 KJ747203 AIG13073.1 ATLV01015210 ATLV01015211 ATLV01015212 ATLV01015213 KE525003 KFB40214.1 GAMC01005659 JAC00897.1 CVRI01000056 CRL01703.1 GFXV01003335 MBW15140.1 CCAG010005258 ADMH02001564 ETN62051.1 DS232425 EDS41623.1 KQ982691 KYQ52204.1 ACZ95173.1 AAAB01008924 EAA44457.4 ABLF02029749 ABLF02029750 ABLF02029753 ABLF02029756 ABLF02029758 CH940655 EDW66442.2 KQ976749 KYN08766.1 CH964291 EDW86521.2 AY050490 AY050491 AY616144 AY616145 AY616146 AY616147 AAF45639.3 AAL12478.1 AAL12479.1 AAN09051.2 AAT40457.1 AAT40458.1 AAT40459.1 AAT40460.1 ACZ95172.1 JXJN01022924 JXJN01022925 GGMS01009222 MBY78425.1 AL031027 CAA19838.1 CM000366 EDX16850.1 GBYB01013238 JAG83005.1 AF381249 AAM46167.1 JXUM01058181 JXUM01058182 KQ561991 KXJ76968.1 KX016789 APF30038.1 KX016785 APF30034.1
AXF48844.1 AGBW02010125 OWR49121.1 KQ459590 KPI97364.1 JTDY01000244 KOB78085.1 NWSH01001177 PCG72244.1 LC017780 BAR64794.1 KJ542735 AJD81619.1 GCVX01000126 JAI18104.1 KQ461190 KPJ07180.1 GDKB01000028 JAP38468.1 KY283581 AST36241.1 KQ971326 KYB28456.1 GBYB01013240 JAG83007.1 KYB28457.1 QOIP01000001 RLU26874.1 KQ981272 KYN43865.1 KQ980765 KYN13357.1 KQ414627 KOC67486.1 KZ288212 PBC32826.1 KK107242 EZA54544.1 JRES01000755 KNC28623.1 KP843222 ALD51358.1 UFQS01002934 UFQT01000673 UFQT01002934 SSX14928.1 SSX26393.1 KJ747199 KJ747200 KJ747202 AIG13069.1 AIG13070.1 AIG13072.1 KJ747210 AIG13080.1 KJ747205 AIG13075.1 KJ747206 AIG13076.1 CH379063 KRT06085.1 CP012528 ALC48300.1 KQ976488 KYM83635.1 CH902622 KPU75185.1 EAL32487.3 KRT06083.1 CH933812 KRG07184.1 KJ747201 AIG13071.1 EDV34526.1 CH477485 EAT40152.1 EDW06207.2 KJ747207 AIG13077.1 KRT06084.1 CH479190 EDW25870.1 CM000162 EDX00911.1 AXCM01010787 AXCM01010788 KJ747204 AIG13074.1 AE014298 AHN59259.1 KJ747208 AIG13078.1 CH954183 EDV45490.1 OUUW01000011 SPP86742.1 KJ747209 AIG13079.1 AY616148 AAF45640.2 AAT40461.1 SPP86744.1 CH480825 EDW43838.1 CH916376 EDV95445.1 SPP86745.1 AY616149 AAT40462.1 ACZ95174.1 CH478081 EAT34043.1 KJ747203 AIG13073.1 ATLV01015210 ATLV01015211 ATLV01015212 ATLV01015213 KE525003 KFB40214.1 GAMC01005659 JAC00897.1 CVRI01000056 CRL01703.1 GFXV01003335 MBW15140.1 CCAG010005258 ADMH02001564 ETN62051.1 DS232425 EDS41623.1 KQ982691 KYQ52204.1 ACZ95173.1 AAAB01008924 EAA44457.4 ABLF02029749 ABLF02029750 ABLF02029753 ABLF02029756 ABLF02029758 CH940655 EDW66442.2 KQ976749 KYN08766.1 CH964291 EDW86521.2 AY050490 AY050491 AY616144 AY616145 AY616146 AY616147 AAF45639.3 AAL12478.1 AAL12479.1 AAN09051.2 AAT40457.1 AAT40458.1 AAT40459.1 AAT40460.1 ACZ95172.1 JXJN01022924 JXJN01022925 GGMS01009222 MBY78425.1 AL031027 CAA19838.1 CM000366 EDX16850.1 GBYB01013238 JAG83005.1 AF381249 AAM46167.1 JXUM01058181 JXUM01058182 KQ561991 KXJ76968.1 KX016789 APF30038.1 KX016785 APF30034.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000037510
UP000218220
UP000053240
+ More
UP000007266 UP000279307 UP000078541 UP000078492 UP000053825 UP000242457 UP000005203 UP000053097 UP000037069 UP000095301 UP000095300 UP000001819 UP000092553 UP000078540 UP000007801 UP000009192 UP000008820 UP000008744 UP000002282 UP000075883 UP000000803 UP000008711 UP000268350 UP000075920 UP000001292 UP000001070 UP000075900 UP000030765 UP000075880 UP000069272 UP000183832 UP000002358 UP000192221 UP000091820 UP000092444 UP000000673 UP000002320 UP000075809 UP000007062 UP000092443 UP000007819 UP000008792 UP000078542 UP000007798 UP000092445 UP000092460 UP000000304 UP000069940 UP000249989
UP000007266 UP000279307 UP000078541 UP000078492 UP000053825 UP000242457 UP000005203 UP000053097 UP000037069 UP000095301 UP000095300 UP000001819 UP000092553 UP000078540 UP000007801 UP000009192 UP000008820 UP000008744 UP000002282 UP000075883 UP000000803 UP000008711 UP000268350 UP000075920 UP000001292 UP000001070 UP000075900 UP000030765 UP000075880 UP000069272 UP000183832 UP000002358 UP000192221 UP000091820 UP000092444 UP000000673 UP000002320 UP000075809 UP000007062 UP000092443 UP000007819 UP000008792 UP000078542 UP000007798 UP000092445 UP000092460 UP000000304 UP000069940 UP000249989
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
A0A2H1VNY7
A0A075TD99
H9JQ85
A0A345BF20
A0A212F5V2
A0A194PX73
+ More
A0A0L7LRJ9 A0A2A4JKX7 A0A0F7QHN7 A0A0B5A3G8 A0A0K8TUD8 A0A194QQE4 A0A0V0J153 A0A223HCZ7 A0A139WKM8 A0A0C9RWB2 A0A139WKV8 A0A3L8E2E5 A0A195FTC7 A0A195DKJ2 A0A0L7R9Q3 A0A2A3ENQ0 A0A088A9N9 A0A026WHI8 A0A0L0C858 A0A0M3SBL9 A0A336MC74 A0A1I8M7N9 A0A1I8PVH3 A0A075Q6F7 A0A075Q6G4 A0A075Q6G2 A0A075Q362 A0A0R3P3I3 A0A0M4F7S4 A0A195BG93 A0A0P8XKB0 Q29J15 A0A1S4FIQ8 A0A0Q9XF30 A0A075Q359 B3MRX1 Q16ZR9 B4L761 A0A075QB19 A0A0R3NYN4 B4GTK0 B4PY38 A0A182MIQ3 A0A075Q109 X2JDW0 A0A075Q438 B3P9J4 A0A3B0K250 A0A075Q116 Q9W581 A0A3B0JXG0 A0A182W628 B4I9F7 B4JXG9 A0A3B0JXF2 Q6IWN7 Q16IF5 A0A182RVV6 A0A075Q434 A0A084VQH0 A0A182IWF8 A0A182FMM9 W8C4A0 A0A1J1IN94 K7J2X0 A0A2H8TMD0 A0A1S4G051 A0A1W4VT10 A0A1W4VSC6 A0A1A9X1D8 A0A1B0GFF3 W5JDA1 B0X6V2 A0A151WWC7 E1JJC6 Q7PHP7 A0A1A9YQ94 J9K6V7 B4MB33 A0A195D7B3 B4NPT1 A0A1B0A0B1 Q8MM14 A0A1B0BZ01 A0A2S2QKY9 O76903 B4R2Z6 A0A0C9QFF6 Q8MX77 A0A182GEK7 A0A1L2YXQ5 A0A1L2YXN9
A0A0L7LRJ9 A0A2A4JKX7 A0A0F7QHN7 A0A0B5A3G8 A0A0K8TUD8 A0A194QQE4 A0A0V0J153 A0A223HCZ7 A0A139WKM8 A0A0C9RWB2 A0A139WKV8 A0A3L8E2E5 A0A195FTC7 A0A195DKJ2 A0A0L7R9Q3 A0A2A3ENQ0 A0A088A9N9 A0A026WHI8 A0A0L0C858 A0A0M3SBL9 A0A336MC74 A0A1I8M7N9 A0A1I8PVH3 A0A075Q6F7 A0A075Q6G4 A0A075Q6G2 A0A075Q362 A0A0R3P3I3 A0A0M4F7S4 A0A195BG93 A0A0P8XKB0 Q29J15 A0A1S4FIQ8 A0A0Q9XF30 A0A075Q359 B3MRX1 Q16ZR9 B4L761 A0A075QB19 A0A0R3NYN4 B4GTK0 B4PY38 A0A182MIQ3 A0A075Q109 X2JDW0 A0A075Q438 B3P9J4 A0A3B0K250 A0A075Q116 Q9W581 A0A3B0JXG0 A0A182W628 B4I9F7 B4JXG9 A0A3B0JXF2 Q6IWN7 Q16IF5 A0A182RVV6 A0A075Q434 A0A084VQH0 A0A182IWF8 A0A182FMM9 W8C4A0 A0A1J1IN94 K7J2X0 A0A2H8TMD0 A0A1S4G051 A0A1W4VT10 A0A1W4VSC6 A0A1A9X1D8 A0A1B0GFF3 W5JDA1 B0X6V2 A0A151WWC7 E1JJC6 Q7PHP7 A0A1A9YQ94 J9K6V7 B4MB33 A0A195D7B3 B4NPT1 A0A1B0A0B1 Q8MM14 A0A1B0BZ01 A0A2S2QKY9 O76903 B4R2Z6 A0A0C9QFF6 Q8MX77 A0A182GEK7 A0A1L2YXQ5 A0A1L2YXN9
PDB
5FXK
E-value=4.8653e-126,
Score=1159
Ontologies
GO
Topology
Subcellular location
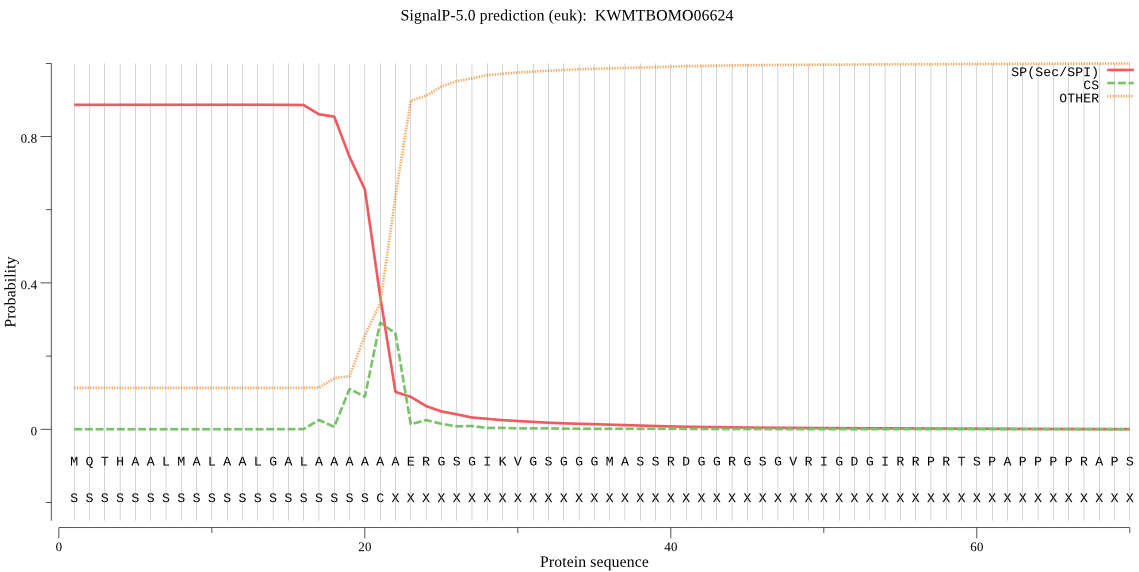
SignalP
Position: 1 - 21,
Likelihood: 0.885746
Length:
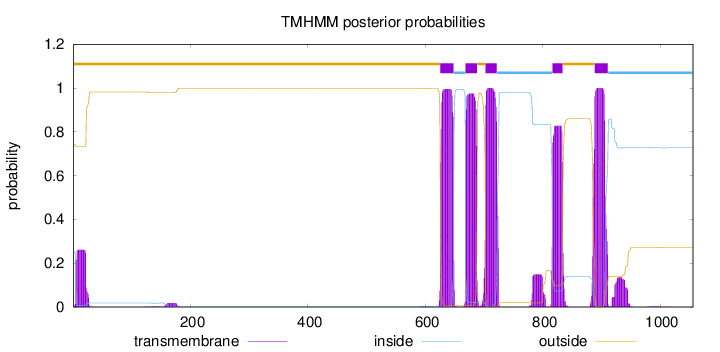
1056
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
111.97612
Exp number, first 60 AAs:
4.94768
Total prob of N-in:
0.25707
outside
1 - 625
TMhelix
626 - 648
inside
649 - 668
TMhelix
669 - 688
outside
689 - 702
TMhelix
703 - 722
inside
723 - 816
TMhelix
817 - 834
outside
835 - 888
TMhelix
889 - 911
inside
912 - 1056
Population Genetic Test Statistics
Pi
169.166465
Theta
161.933405
Tajima's D
0.141218
CLR
0.634602
CSRT
0.406629668516574
Interpretation
Uncertain
