Gene
KWMTBOMO06622 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011786
Annotation
ganglioside-induced_differentiation-associated-protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.625
Sequence
CDS
ATGCACTACGTTCAGAAGTATTTAGAAAAACTACCAATCTCCAAATCAAACATGAAAACTAACGAAGGCAAAAAATGTAGTATTTTTCTTTATTGTAATTATTACAGTTTTTACTCTCAAAAGGTTTTAATGGCTTTATACGAAAAGAATATAGATTTTGAACCCTTAATTGTTGACATAACAAAAGGAGAACAATATTCTTCTTGGTTTCTTGAAATAAATCCTCGTGGAGAAATACCAGTTTTGAAAGTGAAAGATGCGTTAATTCCAGACTCTACTAGGATTTTGGACTATCTGGAATTTTATTTAGACTCTGATTTGGTTCCACTCATAAATGTTTCTAAGGATACAAAAGTGGTAACTACCATAAACAAATTTCGAGAATTGATTGACGCTTTGCCTGCTGGAGTTATAACAGTTGGTTCATTTTTTCATCCACATCTCTGCGGTAGTCCAAAGCTTCCATTTATTTTGCCAGTAAGAGAGATTTTAAAAACTGGTGATTTGAGTAGCTCACAAAACTTGAGAAAGTTGGCGGATGAAAATCCAAAAGCAAAACATATCCTGTTGTACAAAGCTGAGATTCAAGATCGAAAACACGAACTTCTGAGTAATGAAGAGGAATATTTAAAAATTCTAAACATTGTTGACCAAGTTCTGAACCAAATAGAGGAGCAGTTAAAATTACAACCTGTCGGTAATTGGTTATGCTGCGAAAATTTCAGTATTGCAGACATAAATTTGGCTGTCCTCTTGCAACGTCTTTGGGAATTAGGATTAGAAACACGTTTTTGGAGTAACGGAAAGAGACCCTTGATTGAGAGCTATTTTGAACGAGTACGACAAAGAGAATCATTCAAAAATACTATTCCAAATTTGCCACAGCACCTTAAAATGATAGTTACGGCTCCACCTCCAGTTTATGTTGGTGCTGCAGGTGCTGTTTCATTTGCTGTGGTATTTACCGTAGCATACATTTTTAAAAAAATTATTTCGTAA
Protein
MHYVQKYLEKLPISKSNMKTNEGKKCSIFLYCNYYSFYSQKVLMALYEKNIDFEPLIVDITKGEQYSSWFLEINPRGEIPVLKVKDALIPDSTRILDYLEFYLDSDLVPLINVSKDTKVVTTINKFRELIDALPAGVITVGSFFHPHLCGSPKLPFILPVREILKTGDLSSSQNLRKLADENPKAKHILLYKAEIQDRKHELLSNEEEYLKILNIVDQVLNQIEEQLKLQPVGNWLCCENFSIADINLAVLLQRLWELGLETRFWSNGKRPLIESYFERVRQRESFKNTIPNLPQHLKMIVTAPPPVYVGAAGAVSFAVVFTVAYIFKKIIS
Summary
Similarity
Belongs to the GST superfamily.
Uniprot
E7E275
H9JQI0
A0A2H1VQ12
A0A212FJI6
A0A3S2LQR1
A0A194PVE1
+ More
A0A194QNT1 A0A067R0U7 A0A1B6MFW7 A0A1B6LG57 A0A1B6EPU2 A0A1B6GAW8 A0A0T6AU78 A0A1B6IUJ9 A0A2P8YEA1 A0A2R4FXJ4 J3JVD4 N6UDQ8 A0A1Y1NPI9 A0A2J7R8D3 A0A1B6CMX1 D6WFA4 A0A077D023 A0A069DSB0 T1IEE7 A0A0P4VL97 A0A224XGG3 A0A0V0G7P3 A0A023EXY4 A0A182RQY1 A0A2M4A7F8 A0A0A9XRG5 A0A2M3Z744 A0A182PDS8 A0A182WJX9 A0A1Q3FQJ4 W5J854 A0A182GC44 A0A2M4BUS6 A0A182Q9V1 A0A182N825 A0A1S4GRZ1 A0A182XP81 A0A2M4BUK6 A0A182VAC6 Q16VC8 A0A182KNZ9 A0A182IL34 A0A182I1E5 A0A146L7G2 A0A182Y2S2 A0A182SU25 A0A0R1DVR1 B4PIR7 Q9VRJ3 X2JAP0 A0A0K8TIR7 A0A2M4A7Y3 A0A2R7WQE6 A0A0A9Y668 A0A1W4UEH5 M9PBQ8 A0A182TPW1 A0A0A9XY69 A0A0J9RQS1 B4HUD9 B4QRI5 A0A084VW73 B3NG96 A0A0Q5UEJ9 B4N5H8 A0A1L8DAM9 A0A3B0JYP2 A0A182LSI2 A0A034VW07 Q7Q5Y6 W8BAS4 B0WR16 B4H1I7 A0A0R3P2H8 Q29FC3 B4LFQ4 A0A0Q9WRY2 A0A1S4FMR8 Q16E84 B4IX03 A0A0M4EMQ6 B4KWZ7 A0A0Q9XLT9 B3M402 A0A0P8XUY0 A0A0P4W8D0 T1GGF0 A0A182JPU2 A0A0Q5UIH8 A0A1B0CAU6 E0V9E7 A0A336M4W4
A0A194QNT1 A0A067R0U7 A0A1B6MFW7 A0A1B6LG57 A0A1B6EPU2 A0A1B6GAW8 A0A0T6AU78 A0A1B6IUJ9 A0A2P8YEA1 A0A2R4FXJ4 J3JVD4 N6UDQ8 A0A1Y1NPI9 A0A2J7R8D3 A0A1B6CMX1 D6WFA4 A0A077D023 A0A069DSB0 T1IEE7 A0A0P4VL97 A0A224XGG3 A0A0V0G7P3 A0A023EXY4 A0A182RQY1 A0A2M4A7F8 A0A0A9XRG5 A0A2M3Z744 A0A182PDS8 A0A182WJX9 A0A1Q3FQJ4 W5J854 A0A182GC44 A0A2M4BUS6 A0A182Q9V1 A0A182N825 A0A1S4GRZ1 A0A182XP81 A0A2M4BUK6 A0A182VAC6 Q16VC8 A0A182KNZ9 A0A182IL34 A0A182I1E5 A0A146L7G2 A0A182Y2S2 A0A182SU25 A0A0R1DVR1 B4PIR7 Q9VRJ3 X2JAP0 A0A0K8TIR7 A0A2M4A7Y3 A0A2R7WQE6 A0A0A9Y668 A0A1W4UEH5 M9PBQ8 A0A182TPW1 A0A0A9XY69 A0A0J9RQS1 B4HUD9 B4QRI5 A0A084VW73 B3NG96 A0A0Q5UEJ9 B4N5H8 A0A1L8DAM9 A0A3B0JYP2 A0A182LSI2 A0A034VW07 Q7Q5Y6 W8BAS4 B0WR16 B4H1I7 A0A0R3P2H8 Q29FC3 B4LFQ4 A0A0Q9WRY2 A0A1S4FMR8 Q16E84 B4IX03 A0A0M4EMQ6 B4KWZ7 A0A0Q9XLT9 B3M402 A0A0P8XUY0 A0A0P4W8D0 T1GGF0 A0A182JPU2 A0A0Q5UIH8 A0A1B0CAU6 E0V9E7 A0A336M4W4
Pubmed
22316564
19121390
22118469
26354079
24845553
29403074
+ More
22516182 23537049 28004739 18362917 19820115 26334808 27129103 25474469 25401762 20920257 23761445 26483478 12364791 17510324 20966253 26823975 25244985 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 25348373 24495485 15632085 20566863
22516182 23537049 28004739 18362917 19820115 26334808 27129103 25474469 25401762 20920257 23761445 26483478 12364791 17510324 20966253 26823975 25244985 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 25348373 24495485 15632085 20566863
EMBL
HQ601901
ADT82940.1
BABH01012013
ODYU01003588
SOQ42542.1
AGBW02008266
+ More
OWR53892.1 RSAL01000414 RVE41835.1 KQ459590 KPI97361.1 KQ461190 KPJ07183.1 KK853119 KDR11132.1 GEBQ01005188 JAT34789.1 GEBQ01017302 JAT22675.1 GECZ01029817 JAS39952.1 GECZ01010356 JAS59413.1 LJIG01022786 KRT78723.1 GECU01017097 GECU01009073 JAS90609.1 JAS98633.1 PYGN01000666 PSN42578.1 MF034831 AVT42197.1 BT127201 AEE62163.1 APGK01026530 KB740635 KB631992 ENN79840.1 ERL87753.1 GEZM01001139 GEZM01001137 JAV98127.1 NEVH01006722 PNF37103.1 GEDC01022502 JAS14796.1 KQ971319 EFA00477.1 KJ868756 AIL23557.1 GBGD01002273 JAC86616.1 ACPB03014080 ACPB03014081 GDKW01003209 JAI53386.1 GFTR01004851 JAW11575.1 GECL01002658 JAP03466.1 GBBI01004537 JAC14175.1 GGFK01003402 MBW36723.1 GBHO01021388 JAG22216.1 GGFM01003573 MBW24324.1 GFDL01005253 JAV29792.1 ADMH02002104 ETN59045.1 JXUM01009378 JXUM01009379 KQ560282 KXJ83307.1 GGFJ01007623 MBW56764.1 AXCN02000914 AAAB01008960 GGFJ01007625 MBW56766.1 CH477597 EAT38505.1 EJY57813.1 APCN01000054 GDHC01014326 JAQ04303.1 CM000159 KRK01222.1 EDW93487.2 AE014296 AY071643 AAF50802.1 AAL49265.1 AHN57964.1 GBRD01000412 JAG65409.1 GGFK01003554 MBW36875.1 KK855279 PTY21796.1 GBHO01018573 GBHO01018572 GBHO01018571 GBHO01018569 GBRD01000411 GBRD01000410 JAG25031.1 JAG25032.1 JAG25033.1 JAG25035.1 JAG65410.1 AGB94132.1 GBHO01021389 GDHC01001550 JAG22215.1 JAQ17079.1 CM002912 KMY97709.1 CH480817 EDW50560.1 CM000363 EDX09296.1 KMY97708.1 ATLV01017495 KE525170 KFB42217.1 CH954178 EDV50997.1 KQS43617.1 CH964101 EDW79617.1 GFDF01010659 JAV03425.1 OUUW01000012 SPP87164.1 AXCM01000220 GAKP01013271 GAKP01013269 JAC45683.1 EAA10845.3 GAMC01016349 JAB90206.1 DS232049 EDS33114.1 CH479202 EDW30164.1 CH379067 KRT07403.1 EAL31332.2 CH940647 EDW69281.1 KRF84289.1 CH478959 EAT32542.1 CH916366 EDV96309.1 CP012525 ALC43080.1 CH933809 EDW18618.1 KRG06276.1 CH902618 EDV40364.1 KPU78531.1 GDRN01073768 GDRN01073767 JAI63333.1 CAQQ02006538 CAQQ02006539 CAQQ02006540 KQS43616.1 AJWK01004272 DS234992 EEB10003.1 UFQT01000164 SSX21058.1
OWR53892.1 RSAL01000414 RVE41835.1 KQ459590 KPI97361.1 KQ461190 KPJ07183.1 KK853119 KDR11132.1 GEBQ01005188 JAT34789.1 GEBQ01017302 JAT22675.1 GECZ01029817 JAS39952.1 GECZ01010356 JAS59413.1 LJIG01022786 KRT78723.1 GECU01017097 GECU01009073 JAS90609.1 JAS98633.1 PYGN01000666 PSN42578.1 MF034831 AVT42197.1 BT127201 AEE62163.1 APGK01026530 KB740635 KB631992 ENN79840.1 ERL87753.1 GEZM01001139 GEZM01001137 JAV98127.1 NEVH01006722 PNF37103.1 GEDC01022502 JAS14796.1 KQ971319 EFA00477.1 KJ868756 AIL23557.1 GBGD01002273 JAC86616.1 ACPB03014080 ACPB03014081 GDKW01003209 JAI53386.1 GFTR01004851 JAW11575.1 GECL01002658 JAP03466.1 GBBI01004537 JAC14175.1 GGFK01003402 MBW36723.1 GBHO01021388 JAG22216.1 GGFM01003573 MBW24324.1 GFDL01005253 JAV29792.1 ADMH02002104 ETN59045.1 JXUM01009378 JXUM01009379 KQ560282 KXJ83307.1 GGFJ01007623 MBW56764.1 AXCN02000914 AAAB01008960 GGFJ01007625 MBW56766.1 CH477597 EAT38505.1 EJY57813.1 APCN01000054 GDHC01014326 JAQ04303.1 CM000159 KRK01222.1 EDW93487.2 AE014296 AY071643 AAF50802.1 AAL49265.1 AHN57964.1 GBRD01000412 JAG65409.1 GGFK01003554 MBW36875.1 KK855279 PTY21796.1 GBHO01018573 GBHO01018572 GBHO01018571 GBHO01018569 GBRD01000411 GBRD01000410 JAG25031.1 JAG25032.1 JAG25033.1 JAG25035.1 JAG65410.1 AGB94132.1 GBHO01021389 GDHC01001550 JAG22215.1 JAQ17079.1 CM002912 KMY97709.1 CH480817 EDW50560.1 CM000363 EDX09296.1 KMY97708.1 ATLV01017495 KE525170 KFB42217.1 CH954178 EDV50997.1 KQS43617.1 CH964101 EDW79617.1 GFDF01010659 JAV03425.1 OUUW01000012 SPP87164.1 AXCM01000220 GAKP01013271 GAKP01013269 JAC45683.1 EAA10845.3 GAMC01016349 JAB90206.1 DS232049 EDS33114.1 CH479202 EDW30164.1 CH379067 KRT07403.1 EAL31332.2 CH940647 EDW69281.1 KRF84289.1 CH478959 EAT32542.1 CH916366 EDV96309.1 CP012525 ALC43080.1 CH933809 EDW18618.1 KRG06276.1 CH902618 EDV40364.1 KPU78531.1 GDRN01073768 GDRN01073767 JAI63333.1 CAQQ02006538 CAQQ02006539 CAQQ02006540 KQS43616.1 AJWK01004272 DS234992 EEB10003.1 UFQT01000164 SSX21058.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053268
UP000053240
UP000027135
+ More
UP000245037 UP000019118 UP000030742 UP000235965 UP000007266 UP000015103 UP000075900 UP000075885 UP000075920 UP000000673 UP000069940 UP000249989 UP000075886 UP000075884 UP000076407 UP000075903 UP000008820 UP000075882 UP000075880 UP000075840 UP000076408 UP000075901 UP000002282 UP000000803 UP000192221 UP000075902 UP000001292 UP000000304 UP000030765 UP000008711 UP000007798 UP000268350 UP000075883 UP000007062 UP000002320 UP000008744 UP000001819 UP000008792 UP000001070 UP000092553 UP000009192 UP000007801 UP000015102 UP000075881 UP000092461 UP000009046
UP000245037 UP000019118 UP000030742 UP000235965 UP000007266 UP000015103 UP000075900 UP000075885 UP000075920 UP000000673 UP000069940 UP000249989 UP000075886 UP000075884 UP000076407 UP000075903 UP000008820 UP000075882 UP000075880 UP000075840 UP000076408 UP000075901 UP000002282 UP000000803 UP000192221 UP000075902 UP000001292 UP000000304 UP000030765 UP000008711 UP000007798 UP000268350 UP000075883 UP000007062 UP000002320 UP000008744 UP000001819 UP000008792 UP000001070 UP000092553 UP000009192 UP000007801 UP000015102 UP000075881 UP000092461 UP000009046
Interpro
ProteinModelPortal
E7E275
H9JQI0
A0A2H1VQ12
A0A212FJI6
A0A3S2LQR1
A0A194PVE1
+ More
A0A194QNT1 A0A067R0U7 A0A1B6MFW7 A0A1B6LG57 A0A1B6EPU2 A0A1B6GAW8 A0A0T6AU78 A0A1B6IUJ9 A0A2P8YEA1 A0A2R4FXJ4 J3JVD4 N6UDQ8 A0A1Y1NPI9 A0A2J7R8D3 A0A1B6CMX1 D6WFA4 A0A077D023 A0A069DSB0 T1IEE7 A0A0P4VL97 A0A224XGG3 A0A0V0G7P3 A0A023EXY4 A0A182RQY1 A0A2M4A7F8 A0A0A9XRG5 A0A2M3Z744 A0A182PDS8 A0A182WJX9 A0A1Q3FQJ4 W5J854 A0A182GC44 A0A2M4BUS6 A0A182Q9V1 A0A182N825 A0A1S4GRZ1 A0A182XP81 A0A2M4BUK6 A0A182VAC6 Q16VC8 A0A182KNZ9 A0A182IL34 A0A182I1E5 A0A146L7G2 A0A182Y2S2 A0A182SU25 A0A0R1DVR1 B4PIR7 Q9VRJ3 X2JAP0 A0A0K8TIR7 A0A2M4A7Y3 A0A2R7WQE6 A0A0A9Y668 A0A1W4UEH5 M9PBQ8 A0A182TPW1 A0A0A9XY69 A0A0J9RQS1 B4HUD9 B4QRI5 A0A084VW73 B3NG96 A0A0Q5UEJ9 B4N5H8 A0A1L8DAM9 A0A3B0JYP2 A0A182LSI2 A0A034VW07 Q7Q5Y6 W8BAS4 B0WR16 B4H1I7 A0A0R3P2H8 Q29FC3 B4LFQ4 A0A0Q9WRY2 A0A1S4FMR8 Q16E84 B4IX03 A0A0M4EMQ6 B4KWZ7 A0A0Q9XLT9 B3M402 A0A0P8XUY0 A0A0P4W8D0 T1GGF0 A0A182JPU2 A0A0Q5UIH8 A0A1B0CAU6 E0V9E7 A0A336M4W4
A0A194QNT1 A0A067R0U7 A0A1B6MFW7 A0A1B6LG57 A0A1B6EPU2 A0A1B6GAW8 A0A0T6AU78 A0A1B6IUJ9 A0A2P8YEA1 A0A2R4FXJ4 J3JVD4 N6UDQ8 A0A1Y1NPI9 A0A2J7R8D3 A0A1B6CMX1 D6WFA4 A0A077D023 A0A069DSB0 T1IEE7 A0A0P4VL97 A0A224XGG3 A0A0V0G7P3 A0A023EXY4 A0A182RQY1 A0A2M4A7F8 A0A0A9XRG5 A0A2M3Z744 A0A182PDS8 A0A182WJX9 A0A1Q3FQJ4 W5J854 A0A182GC44 A0A2M4BUS6 A0A182Q9V1 A0A182N825 A0A1S4GRZ1 A0A182XP81 A0A2M4BUK6 A0A182VAC6 Q16VC8 A0A182KNZ9 A0A182IL34 A0A182I1E5 A0A146L7G2 A0A182Y2S2 A0A182SU25 A0A0R1DVR1 B4PIR7 Q9VRJ3 X2JAP0 A0A0K8TIR7 A0A2M4A7Y3 A0A2R7WQE6 A0A0A9Y668 A0A1W4UEH5 M9PBQ8 A0A182TPW1 A0A0A9XY69 A0A0J9RQS1 B4HUD9 B4QRI5 A0A084VW73 B3NG96 A0A0Q5UEJ9 B4N5H8 A0A1L8DAM9 A0A3B0JYP2 A0A182LSI2 A0A034VW07 Q7Q5Y6 W8BAS4 B0WR16 B4H1I7 A0A0R3P2H8 Q29FC3 B4LFQ4 A0A0Q9WRY2 A0A1S4FMR8 Q16E84 B4IX03 A0A0M4EMQ6 B4KWZ7 A0A0Q9XLT9 B3M402 A0A0P8XUY0 A0A0P4W8D0 T1GGF0 A0A182JPU2 A0A0Q5UIH8 A0A1B0CAU6 E0V9E7 A0A336M4W4
PDB
4XT0
E-value=4.89348e-10,
Score=154
Ontologies
GO
Topology
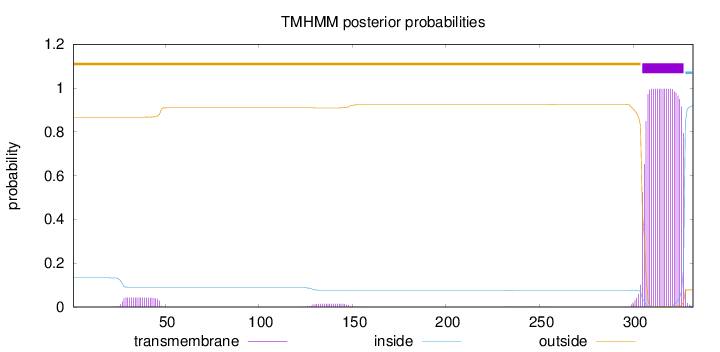
Length:
332
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.11086
Exp number, first 60 AAs:
0.87835
Total prob of N-in:
0.13335
outside
1 - 304
TMhelix
305 - 327
inside
328 - 332
Population Genetic Test Statistics
Pi
190.398845
Theta
168.65021
Tajima's D
0.672198
CLR
0.108565
CSRT
0.560921953902305
Interpretation
Uncertain
